Daumke Lab
Structural Biology of Membrane-Associated Processes
Structural insights into dynamin-mediated endocytosis
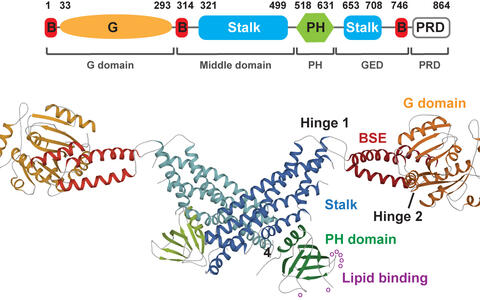
Figure 1: a) Domain architecture of dynamin and b) and crystal structure (pdb 3SNH) of the dynamin dimer.
The multi-domain protein dynamin oligomerizes around the neck of clathrin-coated vesicles into helical filaments which induce membrane scission in a GTPase-dependent fashion. To characterize its molecular mechanisms of action, we determined X-ray structures of a non-oligomerizing, almost full-length dynamin 1 dimer (Faelber et al., Nature, 2011) and of an auto-inhibited dynamin tetramer (Reubold et al., Nature, 2015).
Dynamin comprises a four-domain architecture, including a GTPase domain, a bundle signaling element (BSE), a stalk and a lipid-binding pleckstrin homology (PH) domain (Fig. 1). We showed that the stalk mediates assembly of dynamin into a helical filament. The GTPase domain with the BSE comprises the motor unit of dynamin. GTP binding induces an open BSE conformation while the BSE closes upon GTP hydrolysis. GTPase domains of adjacent filaments dimerize in the GTP-bound form. Dimerization stimulates GTP hydrolysis and the subsequent BSE closure acts as a powerstroke. Based on the structural work from us and others, we suggested a model for dynamin-dependent membrane constriction (Daumke et al., Cell, 2014) (Antonny et al., EMBO J, 2016) (Daumke and Praefcke, Biopolymers, 2016) (Movie 1).
Movie 1: Structural model for dynamin-mediated membrane constriction.
More recently, we biochemically characterized the nucleotide hydrolysis cycle of dynamin using a motor domain construct consisting of the GTPase domain and BSE (Ganichkin et al., PNAS, 2021). We employed bulk FRET analyses to monitor the kinetics of BSE opening in response to GTP binding. Using single molecule FRET analyses in combination with molecular modelling, we determined the force that a single dynamin motor unit generates during one step of GTP hydrolysis. We incorporated the derived rates, forces and geometries into a coarse-grained computational model of the dynamin helix on a membrane tubule. For this, we used a membrane model that follows the energetics of elastic membrane theory (Noel et al., Biophysical Journal, 2019). The dynamin helix was modelled as an elastic filament, and elastic parameters were derived from all atom molecular dynamics simulations.
Movie 2: Physical modelling of dynamin-mediated membrane constriction testing different dissociation rates of the GTPase domain. The dynamin filament in this simulation contains 28 dynamin dimers. The top panel shows the dynamics in the absence of GTP. The other three panels show the first 2 seconds of simulations after the addition of GTP for kdiss = (100, 33, 10) s−1. White beads (GTP-bound monomers or apo in the top panel), cyan beads (GDP-bound monomers), green beads and links (GTP-bound dimers), red beads and links (GDP-bound dimers). Each bead is a dynamin dimer and has two associated MMs. The color of the bead corresponds to only one of the associated MMs, the coloring for the right rung indicates the state of the MMs pointing to the left and for the left rung indicates the state of the MMs pointing to the right.
Our model faithfully reproduced dynamin-mediated membrane constriction via a ratcheting mechanism in realistic time scales and showed that for efficient membrane constriction, many motor domains must cooperate although their movement is not coordinated. The model also allowed us to estimate parameters of the motor cycle that could not be experimentally derived, such as the dissociation rate of GTPase domains upon GTP hydrolysis. Furthermore, we rationalized previous experimental observations, for example, why dynamin-mediated membrane scission takes place at the ends of a dynamin filament. Taken together, our integrated structural biology, experimental and modelling approach led us to derive a holistic mechanistic understanding of the dynamin-mediated ratcheting mechanism as a prerequisite to understand GTPase-driven membrane fission in eukaryotes.
Publications
- Faelber, K., Posor, Y., Gao, S., Held, M., Roske, Y., Schulze, D., Haucke, V., Noe, F., and Daumke, O. (2011). Crystal structure of nucleotide-free dynamin. Nature 477, 556-560. https://doi.org/10.1038/nature10369.
- Reubold, T.F., Faelber, K., Plattner, N., Posor, Y., Ketel, K., Curth, U., Schlegel, J., Anand, R., Manstein, D.J., Noe, F., et al. (2015). Crystal structure of the dynamin tetramer. Nature 525, 404-408. https://doi.org/10.1038/nature14880.
- Daumke, O., Roux, A., and Haucke, V. (2014). BAR domain scaffolds in dynamin-mediated membrane fission. Cell 156, 882-892. https://doi.org/10.1016/j.cell.2014.02.017.
- Antonny, B., Burd, C., De Camilli, P., Chen, E., Daumke, O., Faelber, K., Ford, M., Frolov, V.A., Frost, A., Hinshaw, J.E., et al. (2016). Membrane fission by dynamin: what we know and what we need to know. EMBO J 35, 2270-2284. https://doi.org/10.15252/embj.201694613.
- Daumke, O., and Praefcke, G.J. (2016). Invited review: Mechanisms of GTP hydrolysis and conformational transitions in the dynamin superfamily. Biopolymers 105, 580-593. https://doi.org/10.1002/bip.22855.
- Ganichkin, O.M., Vancraenenbroeck, R., Rosenblum, G., Hofmann, H., Mikhailov, A.S., Daumke, O., and Noel, J.K. (2021). Quantification and demonstration of the collective constriction-by-ratchet mechanism in the dynamin molecular motor. PNAS 118. https://doi.org/10.1073/pnas.2101144118.
- Noel, J.K., Noe, F., Daumke, O., and Mikhailov, A.S. (2019). Polymer-like Model to Study the Dynamics of Dynamin Filaments on Deformable Membrane Tubes. Biophysical journal 117, 1870-1891. https://doi.org/10.1016/j.bpj.2019.09.042.
Researchers in my group
Katja Fälber (Staff Scientist)
Jeff Noel (PostDoc 2015 – 2021)
Oleg Ganichkin (PostDoc 2016-2019)
Song Gao (PhD Student/PostDoc 2007-2013)
Main collaborations
Volker Haucke, FMP Berlin
Frank Noé, FU Berlin
Susanne Eschenburg, Medical University Hannover
Hagen Hoffmann, Weizmann Institute of Science
Alexander S, Mikhailov, Fritz Haber Institute, Berlin
Press releases