Kudryashev Lab
In Situ Structural Biology
Profil
The major focus of the group is on ion channels which open and close selective pores in the membranes of cells allowing ions to pass from one side of the membrane to another. Ions in turn pass signals, regulate voltage across the membrane or initiate signaling cascades. The activation of the pores of ion channels can be caused by multiple factors such as binding ligands to extracellular domains, mechanical pressure or change in voltage across the membrane. Many diseases are associated with dysfunction of ion channels, therefore understanding he details of their activation has a potential to improve the drugs against cardiac and neurological diseases.
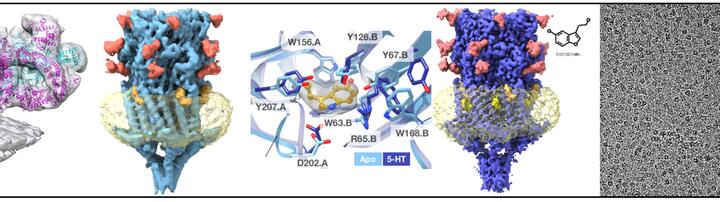
In our work we use cryo electron microscopy as a major tool, it has two flavors. First – so called single particle cryo electron microscopy: proteins are purified, frozen in a thin layer of amorphous ice and are imaged in 2D with an electron microscope. Many identical proteins are imaged in different orientations; this allows determining 3D structure of proteins at resolution where individual amino acids of proteins can be identified. Evolution of hardware and software resulted in the 2017 Nobel Prize in Chemistry to the pioneers of cryo-EM. We use this method to determine structures the serotonin receptor 5HT3r (blue protein on the top image) and showed how binding of serotonin leads to opening of the transmembrane pore and how the lipids such as cholesterol and phospholipids are critically important for protein functionality. We also solved many other protein structures such as the structure of an important protein on the surface of malaria parasites MSP1, which is currently in clinical trials as a vaccine candidate (yellow protein on the right of the top panel).
The second flavor of cryo-EM is cryo electron tomography that allows 3D imaging or unique objects such as viruses, bacteria, cells and even tissues. Repeating membrane proteins can also be identified in such tomograms, mutually aligned using computers and structures can be produced. Structures of membrane proteins without purification, “in situ”, are very useful as they contain physiologically relevant lipids and proteins that are often lost during purification of membrane proteins from membranes using detergents. We used this method to determine the structure of a giant calcium channel ryanodine receptor RyR1 in native membranes (two left panels on the top image). Importantly, may membrane proteins and protein complexes are hard to purify as they disassemble, therefore structural determination by tomography is promising to understand how membrane proteins interact in their native context, “in situ”.
Finally, cryo electron tomograms are very large, noisy and have anisotropic resolution, therefore determining structure of proteins from them is a technical challenge. As a result, the method is only accessible to a limited numbers of researchers with skills in coding and requires a lot of manual interventions. We develop tools to make this process faster and more precise. For this we use high-performance computing with elements of neuronal networks. We realized that in our implementation we can take off a major burden of routine operations from the user, therefore researchers can focus on reaching higher resolution and scientific questions, as opposed to performing routine operations.
If you are interested to join us at any career level for an extended period of time please write me an email to mikhail.kudryashev@mdc-berlin.de
Team
Retreat in Palermo, Italy in Sept 2022. From left to right, standing: Artsemi Yushkevich, Ulja Kravcenko, Misha Kudryashev, Viachaslav Kralin; Vasilii Mikirtumov, Julia Ruta, Xiaofeng Chu, Sabrina Golusik. Bottom row: Dr. Giulia Glorani, Shutian Wang, Rui Li