Membrane couture made of elastic protein bands
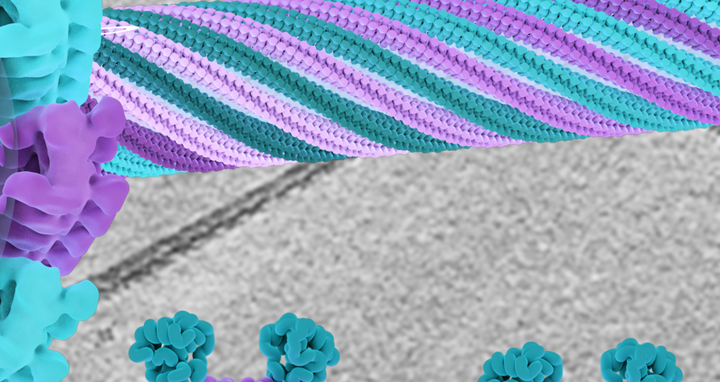
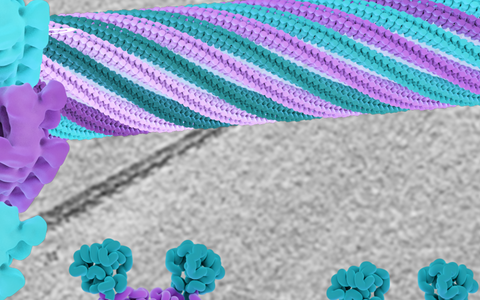
There is constant traffic between our cells and the surrounding extracellular space. While ions and small polar molecules like water pass relatively easily into cells via channel proteins or osmosis, larger “goods” have a more difficult time. Macromolecules, nutrients like fatty acids, and liquid droplets usually enter cells via endocytosis, which is an energy-intensive process. A large patch of the cell membrane folds inward before engulfing the cargo and pinching off completely. These pinched-off pieces of membrane are then further remodeled within the cell into smaller membrane vesicles or tubes. But how does a flat piece of membrane become such a curved vesicle or tube? A protein called EHD4 makes the appropriate “tucks” in the membrane’s “fabric,” much like a tailor would. Professor Oliver Daumke’s team at the Max Delbrück Center has used cryo-electron tomography to see how these tucks are constructed. They are made of a long chain of EHD4 proteins that together form thread-like structures called filaments. The researchers also learned how the necessary curvature in the membrane comes about: it is determined by the curved shape of the EHD4 filament, as they write in the journal “Nature Communications”.
For Daumke, the last author of the paper, this is yet another milestone in what has been his longest research project. From 2004 to 2007, he worked to elucidate the structural workings of EHD proteins as a postdoctoral researcher at the MRC Laboratory of Molecular Biology in Cambridge, UK. Four of these proteins are present in humans. They have been highly conserved in eukaryotes throughout evolution and belong to a family of molecular machines. “Two previous crystal structure analyses showed that EHDs in the absence of membranes occur as dimers,” explains Daumke. “In the first structure, we saw that the two membrane-binding domains of the dimer protrude parallel to each other to form a closed configuration. In the second structure, however, they protrude away from each other in an open configuration.” A key collaborator in the study was first author Dr. Arthur Alves De Melo, who earned his PhD with Daumke back in 2018 on the open EHD4 structure before going on to complete this work.
Ice-cold microscope produces sharp images
We imagine that the filaments function like elastic bands. They apparently adapt dynamically to the curvature of the tubes.
When performing an X-ray structural analysis, it is necessary to purify the proteins and grow high-purity single crystals from them. No one knows whether a protein arranges itself in its crystal form in exactly the same way as it does in a living cell. So it was completely unclear in which of the two conformations the dimer actually exists when bound to the membrane. To find out, the researchers used an advanced microscopy technique called cryo-electron tomography. This emerging technology is suitable for heterogeneous samples such as membrane tubes or even entire sections of cells. Just like in computer tomography – except that here the samples are flash frozen in liquid helium – a sample is imaged from many different angles and the images taken are aligned and merged to create a three-dimensional picture. This makes it possible to reconstruct the architecture of the structures being studied down to the molecular level.
Although the resulting images are not as detailed as those obtained by crystal analysis, in this instance they captured exactly what the researchers wanted to know – namely that the EHD4 dimer assembles in closed form in long filaments on the membrane. “The dimer itself is rigid to a certain extent and forms a curved scaffold, and this curvature is transferred to the flat membrane, which is then remodeled into a tube,” explains Daumke. None of the observed tubes looked alike; some were longer or had a larger diameter. Daumke has a theory about how the different curvatures are generated: “We imagine that the filaments function like elastic bands. They apparently adapt dynamically to the curvature of the tubes. At the beginning, such a filament wraps around a large membrane vesicle like a vine and gradually induces the formation of tubes.”
New microscopy technology on Campus Buch
Despite the comparatively low resolution, Daumke’s team believes this technique has the potential to be groundbreaking. This is because thousands of crystal analyses have already been made of eukaryotic proteins, but so far only about 200 structures have been elucidated and visualized using cryo-electron microscopy; yet these are of very interesting heterogeneous structures that commonly occur within the cell. Such a microscope went into operation last year on Campus Buch. It belongs to Charité – Universitätsmedizin Berlin and is available to all structural biologists in Berlin. The Max Delbrück Center has constructed a building especially for it.
The center of this image shows a structural model of the EHD4 filaments assembling in a helical fashion around a transparent membrane tube. In the background, a cryo-EM micrograph is depicted, which is the basis of the model. At the bottom, two conformations of the EHD4 dimer are displayed which bind to curved (left) and flat (right) membrane surfaces; the left conformation is the one found in the filaments.
“Of course, we would prefer to observe these structures in the cellular environment,” says Daumke. “But until now that was only possible with light microscopy – that is, with much lower magnification.” He now wants to overcome this challenge by combining light and electron microscopy. His team is also currently looking for substances that inhibit EHDs in order to elucidate the functions of this group of proteins at the cellular level.
Text: Catarina Pietschmann
Further information
Literature
Arthur A. Melo et al. (2022): „Cryo-electron tomography reveals structural insights into the membrane remodeling of dynamin-like EHD filaments“. Nature Communications, https://doi.org/10.1038/s41467-022-35164-x
Contacts
Prof. Dr. Oliver Daumke
Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC)
+49 30 9406-3263
oliver.daumke@mdc-berlin.de
Christina Anders
Editor, Communications Department
Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC)
+49 30 9406-2118
christina.anders@mdc-berlin.de or presse@mdc-berlin.de
- Max Delbrück Center
-
The Max Delbrück Center for Molecular Medicine in the Helmholtz Association (Max Delbrück Center) is one of the world’s leading biomedical research institutions. Max Delbrück, a Berlin native, was a Nobel laureate and one of the founders of molecular biology. At the locations in Berlin-Buch and Mitte, researchers from some 70 countries study human biology – investigating the foundations of life from its most elementary building blocks to systems-wide mechanisms. By understanding what regulates or disrupts the dynamic equilibrium of a cell, an organ, or the entire body, we can prevent diseases, diagnose them earlier, and stop their progression with tailored therapies. Patients should be able to benefit as soon as possible from basic research discoveries. This is why the Max Delbrück Center supports spin-off creation and participates in collaborative networks. It works in close partnership with Charité – Universitätsmedizin Berlin in the jointly-run Experimental and Clinical Research Center (ECRC), the Berlin Institute of Health (BIH) at Charité, and the German Center for Cardiovascular Research (DZHK). Founded in 1992, the Max Delbrück Center today employs 1,800 people and is 90 percent funded by the German federal government and 10 percent by the State of Berlin.