Treier Lab
Genetics of Metabolic and Reproductive Disorders
Profile
And which role do transcription factors like GLIS3, GLIS2, BSX, SALL4 and FOXL2 play in the context of Diabetes, CardioRenal Syndrome, Obesity, Sleep Disorders or Sexual Reproduction?
Our group is studying the various aspects of mammalian physiology and homeostasis, from the single cell stage to the complex interplay between organs. With a plethora of self-created mouse models for human diseases complemented by latest technologies we are dissecting highly complex physiological processes at the organismal level.
For further information please visit our research section.
If you get inspired and want to join us in our quest, check out the job options or contact us directly!
Team
Research
Thereto the influence of certain transcription factors on physiological processes, that are involved in diseases like diabetes, obesity and sleep disorders, is investigated in detail by modern techniques for in vivo recording of metabolic parameters and whole tissue imaging.
These are the fields we are currently working on:
- CNS Regulation of Metabolism
-
BSX and its role in obesity and sleep disorders
-
In previous studies we have identified the brain-specific homeobox transcription factor BSX as crucial factor for the transcriptional regulation of the two key orexigenic peptides NPY and AgRP in neurons and for physical activity (Sakkou et al, 2007).
Currently, we are characterizing the neuronal circuits of the CNS that are involved in the dysregulation of physical activity and sleep, both, contributing to the development of obesity in humans.
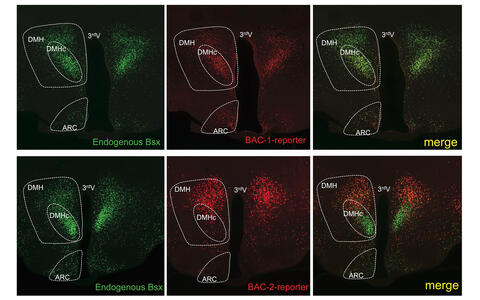
Identification of neuronal enhancer elements employing BAC (Bacterial Artifical Chromosome) mouse transgenesis. Shown is the endogenous expression pattern of BSX in the mouse hypothalamus visualized by Histone2BGFP expression with superimposed expression of BAC reporters (Histone2B-RFP) harboring the BSX locus. Note, the enhancer element for the DMHc (compact region of the Dorsomedial hypothalamus) is missing on the BAC-2 reporter (ARC = arcuate nucleus).
In this project modern transgenic and DREADD technologies are used to silence or activate defined neuronal populations followed by in vivo metabolic phenotyping to determine their influence on the regulation of locomotion behaviour and sleep patterns. With a combination of tissue clearing and light sheet microscopy we’ve started to deconstruct the BSX hypothalamic network. Resulting image acquisitions are terabytes in size and consist of many large, unaligned image tiles that suffer from optical distortions. We developed in cooperation with the Stephan Preibisch Lab an ImageJ package called "BigStitcher" (pre-published here), for adequate analysis of data obtained from light sheet microscopy.
3D Visualisation of BSX-lineage neurones in the mouse hypothalamus using tissue clearing techniques.
3D Visualisation of BSX-lineage neurones in the mouse hypothalamus using tissue clearing techniques
Projections of BSX-expressing neurons throughout the whole mouse brain
Projections of BSX-expressing neurons throughout the whole mouse brain
- Gene-Nutrient Interaction
-
Genetic dissection of CardioRenal Syndrome and Diabetes
-
GLIS2
Our work on GLIS transcription regulators has led to the discovery that mutations in GLIS2, a zinc finger transcriptional regulator, cause nephronophthisis – a progressive chronic kidney disease - in mice and man (Attanatiios et al, 2007).
Employing FGWAS (FunctionalGenomeWideAssociationStudies) and CRISPR/Cas9 transgenic technology, we’re screening for novel genetic mutations which, in combination with a loss of GLIS2 function, induce the so called CardioRenal Syndrome - a subclass of cardiovascular diseases in which heart damage is directly elicited by kidney degradation. This approach might lead to novel targets for therapeutic interventions against CRS.
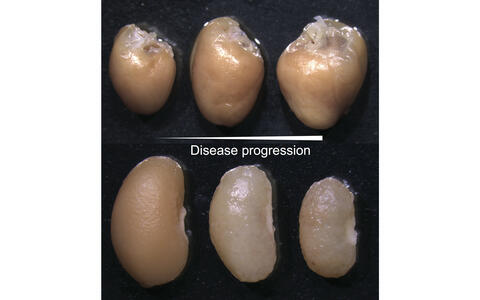
Disease modeling of the CardioRenal syndrome in mice. The degeneration of the kidney overtime is mirrored by increased hyperplasia of the heart (heart top row, kidney below at different time points of disease progression).
GLIS3
According to GWAS, another GLIS family member, GLIS 3, is associated with all forms of diabetes, namely monogenetic, type 1 and type 2 diabetes. GLIS3 mutations are known to cause a multi-organ failure syndrome in humans and mice. In line with this, GLIS3 knock out mice develop diabetes. Over the last years we performed comprehensive studies to analyse the role of GLIS3 in beta-cells in the context of gene-nutrition interaction.
- Stem cells, Plasticity and Regeneration
-
Epigenetic regulation of cell type determination
-
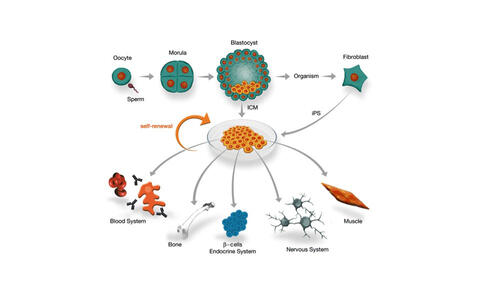
Stem/progenitor cell populations constitute the basic building units from which organs and whole organisms are created.
We identified the transcriptional regulator SALL4 as a key player that is required to maintain the pluripotency state of embryonic stem cells. SALL4 is highly expressed in the inner cell mass (ICM) of the blastocyst that gives rise to the embryo and the primitive endoderm.
We employ omics technologies to understand the regulation and function of this central player in stem cell biology. Using a biotin/streptavidin system we have unraveled the SALL4 protein complex and its protein interaction network in embryonic stem cells. In addition, we have identified the chromosomal localization pattern of these proteins by chromatin immunoprecipitation in combination with high-throughput parallel sequencing (ChIP-Seq).
Currently, we determine the epigenetic alterations that result from changes of these protein complex activities upon growth hormone factor signaling.
This work was part of the Priority Program SPP 1356 "Pluripotency and Cellular Reprogramming" of the German Research Foundation.
- Sexual reproduction
-
From Minnie to Mickey
-
XX = girl and XY = boy
That's what we learnt in school, right? However, there is more to gender phenotype than meets the eye.
We have recently uncovered the molecular mechanism underlying sexual maintenance in mammals revealing an unexpected and fascinating plasticity through a Yin and Yang relationship between two genes, whose expression is mutually exclusive. These two genes are called FOXL2 and SOX9, both transcriptional regulators.
While SOX9 protein expression is abundant in males at all stages of development, it has to be continuously suppressed in adult females to stabilize the female phenotype and to prevent trans-differentiation of the ovaries into testes (see Uhlenhaut et al, Cell 2009).

Technologies
Some of the state-of-the-art techniques used in our lab involve:
- Generation of transgenic mouse models by CRISPR/CAS 9 and BAC Transgenic Technology and laser assisted micromanipulation of mouse embryos (Watch the video!)
- In vivo biotinylation to study protein function
- Tissue Clearing (CLARITY) and Light Sheet Microscopy
- Chemogenetic activation of neurons by DREADDs
- Metabolic Phenotyping
Publications
Selected Publications
Uhlenhaut N, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, Treier AC, Klugmann C, Klasen C, Holter N, Riethmacher D, Schütz G, Cooney A, Lovell-Badge R & Treier M (2009). Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139:1130-1142.
Attanasio M, Uhlenhaut NH, Sousa V, O’Toole JF, Otto E, Anlag K, Klugmann C, Treier AC, Helou J, Sayer JA, Seelow D, Nürnberg G, Becker C, Chudley AE, Nürnberg P, Hildebrandt F & Treier M (2007) Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nature Genetics 39:1018-1024.
Sakkou M, Wiedmer, P, Anlag, K, Hamm, A, Seuntjens, E, Ettwiller, L,Tschop, M & Treier M (2007). A Role for Brain-Specific Homeobox Factor Bsx in the Control of Hyperphagia and Locomotory Behavior. Cell Metabolism 5:450-463
Elling, U., Klasen, C., Eisenberger, T., Anlag, K. & Treier, M (2006). Murine inner cell mass derived lineages depend on Sall4 function. PNAS 103(44), 16319-24.
Schmidt D, Ovitt CE, Anlag K, Fehsenfeld S, Gredsted L, Treier AC, Treier M (2004). The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 131(4):933-42.
Treier M, Gleiberman AS, O'Connell SM, Szeto DP, McMahon JA, McMahon AP, Rosenfeld MG (1998). Multistep signaling requirements for pituitary organogenesis in vivo. Genes & Development 12(11):1691-704.
Treier M, Staszewski LM, Bohmann D (1994). Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78(5): 787-98