A control switch for genes
The bacterial operon has fascinated evolutionary developmental biologists (referred to as evo-devos) ever since the French scientists François Jacob, Jaques Lucien Monod and André Lwoff devised a simple model to explain its workings. The lac operon they described is considered one of the most important starting points for research into gene regulation. For this work they were awarded the 1965 Nobel Prize in Physiology or Medicine.
Dr. Darío G. Lupiáñez, lead organizer of the EMBO workshop
In his keynote lecture at the European Molecular Biology Organization (EMBO) virtual workshop “The evolution of animal genomes,” Michael Levine, a professor of molecular biology and director of the Lewis-Sigler Institute for Integrative Genomis at Princeton University, will examine how the bacterial operon helped to advance evo-devo research and continues to influence it today. The workshop will take place from September 13 to 17, 2021. International experts will discuss how the diversity of life came about and how cutting-edge technologies and techniques – like long-range genome sequencing or chromosome conformation capture methods (3C), which can identify regulatory interactions across the genome – are helping to decipher the genetics of evolution.
Dr. Darío G. Lupiáñez, who heads the Epigenetics and Sex Development Lab at the Berlin Institute for Medical Systems Biology (BIMSB) of the Max Delbrück Center for Molecular Biology in the Helmholtz Association (MDC), is the main organizer of the workshop, to which EMBO is providing funding of €32,600.
- In memory of José Luis Gómez-Skarmeta
-
-
“We are organizing this EMBO workshop in memory of our co-organizer José Luis Gómez-Skarmeta, a distinguished evo-devo researcher and close friend,” says Lupiáñez. Gómez-Skarmeta, who passed away during workshop preparations, pioneered the study of how the non-coding genome regulates complex gene expression patterns and how changes in such sequences can drive evolutionary adaptation. His work contributed greatly to our understanding of gene regulation.
The most elegant Nobel Prize of all time
Professor Levine, what exactly is the bacterial operon?
Michael Levine: The operon is a switch that turns genes on and off. The French scientists François Jacob, Jaques Lucien Monod and André Lwoff devised an incredibly simple model for this phenomenon in the '50s and early '60s. It is based on Escherichia coli mutants whose lactose metabolism is impaired, giving it the name lactose operon, or lac operon for short. When lactose is added to a bacterial culture, enzymes are activated that transport the lactose into the cell and metabolize it there to produce energy. In 1965 this work won the Nobel Prize in Physiology or Medicine – one of the most elegant Nobel Prizes ever awarded!
Elegant in what way?
The lac operon mechanism is simply beautiful by nature.
The lac operon mechanism is simply beautiful by nature. The fact that genes control enzyme synthesis, and thus the basic prerequisite for transcribing genetic information, was a big surprise at the time. Scientists had previously assumed that the enzymes were present in the bacterial cell at the outset, and that they were waiting, so to speak, until they could metabolize lactose. But no, the moment lactose appears, a switch has to be flipped in the DNA before the enzymes required for this metabolism can be formed. That’s exactly what happens during transcription: The cell receives a signal, such as a hormone or a stimulus, and the genes are turned on or not. In bacteria and most other unicellular organisms this occurs directly at the promoter. The promoter is a short segment of the operon that serves as an initiation site for RNA polymerase. RNA polymerase is the enzyme required for transcription – the process of reading genetic information. Close to the promoter is the operator. The operator binds to to proteins that can either stimulate or block the promoter. These are therefore also called activators and repressors. If the operator binds to a repressor, the polymerase pathway becomes blocked – resulting in no transcription, no gene expression and no gene activity. Activators, on the other hand, turn off repressors. When this occurs, the polymerase not only binds to the promoter but also transcribes and activates the gene so that the appropriate proteins are produced. Some 99 percent of all gene regulation in nature works according to this very simple yet so elegant principle.
In complex animals, however, genes are regulated by enhancers. These enhancers, also known as transcription enhancers, are located very far away from genes. They don’t appear to interact directly with one another, but often communicate across several thousand DNA base pairs.
The operon is hiding in plain sight
So the bacterial operon has disappeared over the course of evolution?
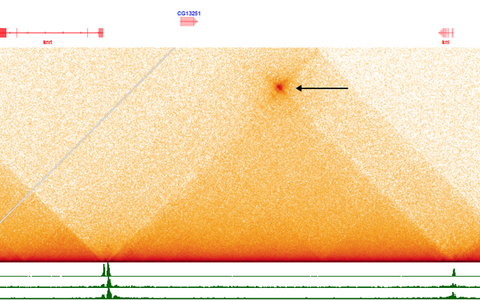
On superficial examination it does indeed look as if this wonderful mechanism has been lost. But with the help of long-range sequencing, two postdoctoral researchers in my group, Joao Raimundo and Xiao Li, have discovered that enhancers are physically connected to promoters as a result of DNA folding. There are also points of contact between promoters of genes that are up to 250,000 nucleotides apart. So, an enhancer can regulate multiple genes at once by turning their promoters on or off – just like the bacterial operon does. We call this switch a “topological operon” or “toperon.” The bacterial operon has therefore not disappeared in higher organisms – it just does its work hiding in plain sight. We believe that about 10 percent of genes are regulated in this way. With the help of genome analysis, we can now identify which genes these are.
Traced with the help of long-range sequencing: the "topological operon".
It all comes down to transcription
Why are you interested in this?
Many researchers are searching for an explanation for the diverse forms of life. I think the answer lies in the mechanisms of gene regulation: It all comes down to transcription. That’s why I want to unlock its secrets.
What questions must still be answered?
The big challenge for evolutionary and developmental biology lies in understanding the folding and dynamics in gene activity. To do this we need to conduct a precise analysis of the genome – not only in terms of its spatial structure, but also how it changes over time.
Deciphering evolution with omics technologies
What role do technologies such as genome sequencing or single-cell analysis play in this respect?
We will understand (...) why elephants have a trunk and humans have five fingers.
Thanks to these technologies, we are currently experiencing an incredibly exciting time. Single-cell technologies make it possible to do things like identify all expressed RNAs, as well as the all active and inactive genes in a cell. They allow us to compare different cell types, helping us understand how they developed during evolution. It is even conceivable that we will be able to reconstruct the ancestor animal with its 20 or 30 different cell types. And we will understand how each of these cell types evolved into hundreds of cell types in humans. Or why elephants have a trunk and humans have five fingers. Once we have understood gene regulation thanks to these technologies, we will also know how different organisms evolved.
Jana Ehrhardt-Joswig conducted the interview.