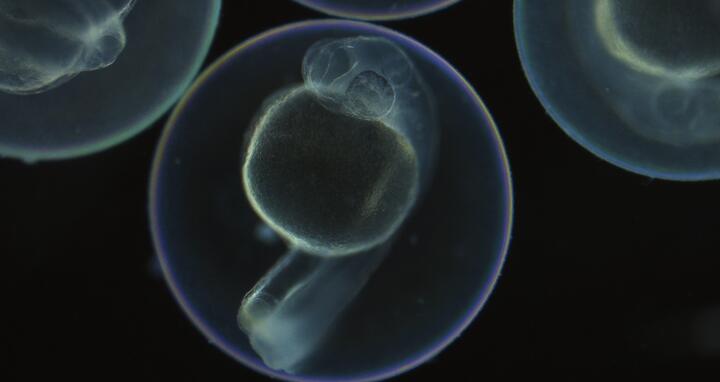
Developmental scars
Where do these different cell types come from?
Every biology textbook states that cells are life’s building blocks. But researchers are only now beginning to understand their diversity. Technologies such as RNA sequencing are revealing which genes are expressed in each individual cell. All cells can then be arranged systematically using similar expression profiles. “Whenever we use such a technology to examine an organ or an organism, we find not only familiar cell types, but also unknown and rare ones,” says Dr. Jan Philipp Junker, head of the Quantitative Developmental Biology research group at the Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC). “The next question is obvious: Where do these different types come from?” Junker’s team is now publishing a paper in the journal Nature Biotechnology that describes a technique called LINNAEUS, which enables researchers to determine the cell type as well as the lineage of each cell.
“We want to understand the flexibility with which organisms develop,” Junker says. If damage occurs during embryonic development, for example, due to a mutation or environmental influences, repair mechanisms ensure that the animal later appears healthy. Only the lineage of each cell can tell the true story – the extent of the damage and the repair mechanism. Even the heart of an adult zebrafish can regenerate after injury. “Is this a biological development process repeating itself or is something new going on?” Junker wonders. “And do cells change and take on other tasks?” Yet in other cases, a missing cell type is responsible for causing a particular disease. In the future, researchers will be able to use the lineage trees of all cells traceable with LINNAEUS to formulate new hypotheses regarding questions such as these.
Rapid patching causes accidental scarring
The technique is based on scars in the DNA, which taken together work like a bar code from which each cell’s lineage can be determined. While the zebrafish embryos are still at the single-cell stage, Junker’s team injects the CRISPR-Cas9 system. Over the next eight hours, Cas9 repeatedly cut a sequence that the fish will certainly never need: the gene for red fluorescent protein (RFP). The embryo’s red glow gradually fades, and thousands of different scars form on the DNA wounds. “CRISPR always makes a cut at an exact point. But the cells have no more than 15 minutes to repair before the next cell division takes place,” Junker says. “Patching must be done quickly, so the chromosome pieces are sort of glued together. This is where mistakes happen. The scars in the DNA are of random length and their exact position also varies.” Daughter cells inherit the genetic scars during cell division. Cells originating from a common ancestor can thus be identified by their genetic scars.
Technologies such as RNA sequencing show which genes are translated in each individual cell. Using similar expression profiles, they were sorted by colour. The black lines symbolize a common origin of the cells. From these data, the researchers traced the cell lineages.
While single-cell RNA sequencing maps thousands of cells by cell type, the scars show millions of connections between the cells. Reconstructing lineage trees from this tangle of data presented a variety of challenges. Some scars are especially likely to occur. “This is dangerous, because if the same scar sequence is created in both heart and brain cells, one might mistakenly assume that they have a common ancestor,” Junker says. So we had to know which sequences we couldn’t trust and filter them out.” Additionally, not all scars in a cell can be found, says bioinformatician Bastiaan Spanjaard, one of the study’s lead authors. “So we have developed a method capable of bridging gaps in the data that enables us to construct lineage trees.”
Zooming in on the data set
The end results are lineage trees with colorful pie charts where the branches divide. Each division is a scar, and each color on the pie chart shows in which cell type it occurs. Researchers can zoom in on this compact representation of an extremely large data set in as much detail as they want.
The researchers were able to observe under the microscope that CRISPR/Cas9 did its work and produced the scars in the genome - because then the red glow of the fish embryos faded.
“In the heart, for instance, there are two cell types that are barely distinguishable. But the lineage trees show that their development branches off in different directions very early on,” says Junker. “We next want to see where these cell types occur in fish hearts. This often provides a first indication of what function they serve.” His lab is continuing to use the zebrafish as a model organism, but Junker also sees great potential in applying the technique to human organoids. This could ultimately help us to understand which mutations in a patient cause permanent damage to cell lineage trees.
Further information
Reference
Bastiaan Spanjaard et al (2018): "Simultaneous lineage tracing and cell-type identification using CRISPR–Cas9-induced genetic scars“, Nature Biotechnology. Advance Online Publication 09.04.2018, doi:10.1038/nbt.4124.
Abstract: A key goal of developmental biology is to understand how a single cell is transformed into a full-grown organism comprising many different cell types. Single-cell RNA-sequencing (scRNA-seq) is commonly used to identify cell types in a tissue or organ. However, organizing the resulting taxonomy of cell types into lineage trees to understand the developmental origin of cells remains challenging. Here we present LINNAEUS (lineage tracing by nuclease-activated editing of ubiquitous sequences) - a strategy for simultaneous lineage tracing and transcriptome profiling in thousands of single cells. By combining scRNA-seq with computational analysis of lineage barcodes, generated by genome editing of transgenic reporter genes, we reconstruct developmental lineage trees in zebrafish larvae, and in heart, liver, pancreas, and telencephalon of adult fish. LINNAEUS provides a systematic approach for tracing the origin of novel cell types, or known cell types under different conditions.
- The Max Delbrück Center for Molecular Medicine (MDC)
-
The Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC) is one of the world’s leading biomedical research institutions. Max Delbrück, a Berlin native, was a Nobel laureate and one of the founders of molecular biology. At the MDC’s locations in Berlin-Buch and Mitte, researchers from some 60 countries analyze the human system – investigating the biological foundations of life from its most elementary building blocks to systems-wide mechanisms. By understanding what regulates or disrupts the dynamic equilibrium in a cell, an organ, or the entire body, we can prevent diseases, diagnose them earlier, and stop their progression with tailored therapies. Patients should benefit as soon as possible from basic research discoveries. The MDC therefore supports spin-off creation and participates in collaborative networks. It works in close partnership with Charité – Universitätsmedizin Berlin in the jointly run Experimental and Clinical Research Center (ECRC), the Berlin Institute of Health (BIH) at Charité, and the German Center for Cardiovascular Research (DZHK). Founded in 1992, the MDC today employs 1,600 people and is funded 90 percent by the German federal government and 10 percent by the State of Berlin.
Contacts for the media
Dr. Jan Philipp Junker
Max Delbrück Center for Molecular Medicine in the Helmholtz Association
The Berlin Institute for Medical Systems Biology
Group Leader "Quantitative Developmental Biology“
JanPhilipp.Junker@mdc-berlin.de
Jana Schlütter
Max Delbrück Center for Molecular Medicine in the Helmholtz Association
Communications department
Phone: +49 30 9406 2121
jana.schluetter@mdc-berlin.de
Main image
LINNAEUS makes it possible to trace the origin of each cell of a zebrafish.