Daumke Lab
Structural Biology of Membrane-Associated Processes
Structural basis for mitochondrial membrane remodeling
Another focus of the group is the structural and functional characterization of mitochondrial membrane remodeling events. Mitochondria are highly dynamic structures with an outer and an inner membrane, which undergo continuous scission and fusion reactions for the maintenance of respiratory and metabolic activity and for preservation of mitochondrial DNA. Malfunction of mitochondrial dynamics has been linked to numerous pathologies, such as neurodegenerative diseases, cardiomyopathy and cancer.
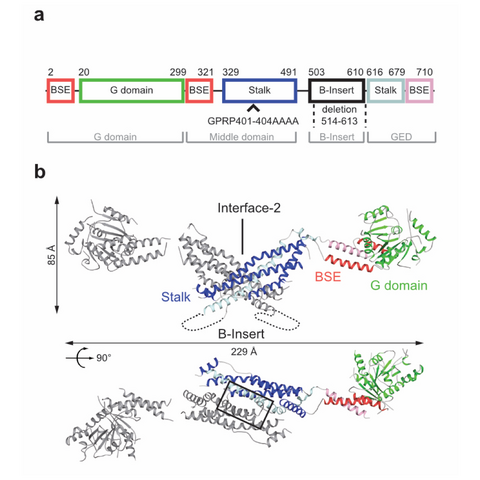
Figure 1: Structural insights into Drp1. Domain architecture (a) and structure (b) of the Drp1 dimer (pdb 4BEJ).
DNM1L mediates fission of mitochondria and peroxisomes, and its dysfunction has been implicated in several neurological disorders, such as Alzheimer's, Parkinson's and Huntington's disease. To study the molecular basis of mitochondrial remodeling, we determined the crystal structure of DNM1L (Frohlich et al., EMBO J, 2013). Similar to dynamin, it is comprised of a GTPase domain, a BSE and a stalk (Fig. 1). DNM1L assembled via a central stalk interface, and mutations in this interface disrupted dimerization and interfered with membrane binding and mitochondrial targeting. Two sequence stretches at the tip of the stalk were shown to be required for ordered assembly of DNM1L on membranes and its function in mitochondrial fission.
Dynamin-like OPA1 (optical atrophy 1) in mammals and its homologue Mgm1 in yeast localize to the intermembrane space and have functions in mediating fusion of the inner mitochondrial membranes and in maintaining crista shape.
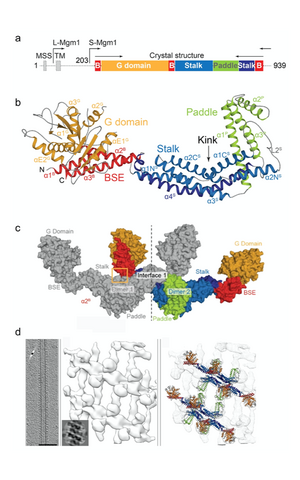
Figure 2: Domain architecture (a) and crystal structure (b) of Mgm1. In the crystals, Mgm1 assembled into a tetramer via conserved interfaces (c). The tetramer was fitted into a low resolution cryo-electron tomography reconstruction of Mgm1 assembled on a lipid tube (d).
We revealed the crystal structure of Mgm1 from a thermophilic yeast (Faelber et al., Nature, 2019) (Fig. 2). Similar to dynamin, Mgm1 consists of a GTPase domain, a BSE, a stalk. Instead of the PH domain, a paddle domain mediates membrane-binding. Biochemical and cell-based experiments demonstrated that the Mgm1 stalk facilitates the assembly of bent tetramers into helical filaments. Electron cryo-tomography studies of Mgm1-decorated lipid tubes revealed the molecular basis of how Mgm1 tetramers assemble at the outside and inside of membrane tubes into membrane remodeling filaments. Fluorescence microscopy experiments on reconstituted membrane tubes confirmed a preference of Mgm1 to bind to the inside of membrane tubes, reflecting the membrane geometry in cristae. Our findings explained how Mgm1 filaments dynamically remodel the mitochondrial inner membrane and led to models of how Mgm1 can maintain crista shape and mediate mitochondrial inner membrane fusion.
More recently, we characterized the molecular function of the conserved, multi-subunit mitochondrial contact site and cristae organizing system (MICOS) complex. MICOS localizes to crista junctions (CJ), which are the tubular openings of the cristae of a defined size. Depletion of certain MICOS subunits leads to loss of cristae junctions, enlarged cristae membranes and impaired mitochondrial energy production. Previous biochemical studies showed that the largest MICOS subunit, Mic60, forms a functional subcomplex with Mic19, but the exact role of this subcomplex was unclear.
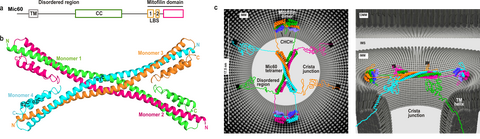
We initially identified a peripheral membrane-binding site in the C-terminal mitofilin-domain of Mic60 (Hessenberger et al., Nat Commun, 2017) To provide first structural insights into CJ formation, we determined the structure of the central coiled-coil (CC) domain of Mic60, which forms an anti-parallel, elongated, bow tie-shaped tetrameric assembly via an evolutionary conserved interface (Bock-Bierbaum et al., Sci Adv, 2022) (Fig. 3). We showed that Mic60 in isolation is mostly in a dimeric state, while interaction with Mic19 promotes its tetrameric assembly via the identified assembly interface. We also showed that the tetrameric interface is crucial for CJ formation in yeast.
Figure 3. Mic60 coiled-coil domain forms an antiparallel tetramer. a) Domain architecture of Mic60. TM: transmembrane helix, CC: coiled-coil, LBS 1+2: lipid binding site. b) Cartoon representation of the Mic60 coiled-coil domain 7PUZ. N- and C-termini of each monomer are labelled. c) Proposed architecture of the Mic60-Mic19 complex at CJs from two views. Each monomer has a different color. Regions not determined by X-ray crystallography are modelled as unstructured elements. OMM/IMM – outer/inner mitochondrial membrane, IMS – intermembrane space. © for Fig. 3c: Erik Werner, RNS Berlin.
Furthermore, we determined the crystal structure of the C-terminal Mic60 mitofilin domain in complex with the CHCH domain of Mic19 (Bock-Bierbaum et al., Sci Adv, 2022). This structure clarified the molecular interaction of Mic60 and Mic19. Furthermore, the structure showed that the mitofilin domain forms a domain-swapped dimer with a crescent-shaped membrane-binding site of convex curvature, which is tailored to interact with the curved membrane at CJ. Combining these data, we suggested a model in which the Mic60-Mic19 subcomplex traverses CJs and functions as a molecular strut that controls CJ architecture and function (Fig. 3). It may also act as a diffusion barrier controlling passage of proteins and metabolites into the cristae space.
Publications
- Frohlich, C., Grabiger, S., Schwefel, D., Faelber, K., Rosenbaum, E., Mears, J., Rocks, O., and Daumke, O. (2013). Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. The EMBO Journal 32, 1280-1292. https://doi.org/10.1038/emboj.2013.74.
- Faelber, K., Dietrich, L., Noel, J.K., Wollweber, F., Pfitzner, A.K., Muhleip, A., Sanchez, R., Kudryashev, M., Chiaruttini, N., Lilie, H., et al. (2019). Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1. Nature 571, 429-433. https://doi.org/10.1038/s41586-019-1372-3.
- Hessenberger, M., Zerbes, R.M., Rampelt, H., Kunz, S., Xavier, A.H., Purfurst, B., Lilie, H., Pfanner, N., van der Laan, M., and Daumke, O. (2017). Regulated membrane remodeling by Mic60 controls formation of mitochondrial crista junctions. Nat Commun 8, 15258. https://doi.org/10.1038/ncomms15258.
- Bock-Bierbaum, T., Funck, K., Wollweber, F., Lisicki, E., von der Malsburg, K., von der Malsburg, A., Laborenz, J., Noel, J.K., Hessenberger, M., Jungbluth, S., et al. (2022). Structural insights into crista junction formation by the Mic60-Mic19 complex. Sci Adv 8, eabo4946. https://doi.org/10.1126/sciadv.abo4946.
Researchers in my group
Tobias Bock-Bierbaum (PostDoc)
Evangelia Nathanail (Doctoral student)
Ashwin Natarajan (PostDoc)
Katja Fälber (Staff Scientist)
Kathrin Funck (Doctoral student, 2017 – 2022)
Manuel Hessenberger (Doctoral student 2013 – 2018)
Chris Fröhlich (Doctoral student 2009 – 2013)
Main collaborators
Martin van der Laan/University of Homburg
Werner Kühlbrandt/MPI Frankfurt
Aurélien Roux/University of Geneva.