Enigmatic loop in the genome
Balancing a stack of paper, a laptop, notebooks, a cell phone and a diminutive coffee cup, Professor Ana Pombo hurries through the corridors of her laboratory. Her office is a glass box in the center of the lab. But it is already occupied by two colleagues, who are busy looking at a screen full of data. “I’m sorry, we have a guest today,” says Pombo, and then takes a seat in the seminar room next door. She dials the number of her colleague Dr. Konstantina Skourti-Stathaki on her phone and puts the device on the edge of her laptop screen.
Unchartered waters
The regulation of these genes is much more complex than we thought, and unexpectedly multilayered.
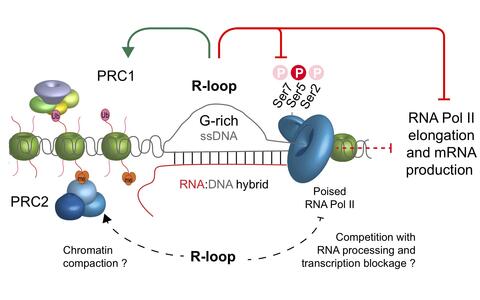
Skourti-Stathaki was also a guest in Ana Pombo’s group recently. She joined her to venture far into unknown research territory, investigating developmental genes that decide whether the cells of an embryo develop into the differentiated cells of the muscle, liver or nervous system. “The regulation of these genes is much more complex than we thought, and unexpectedly multilayered,” says Skourti-Stathaki. Especially important are the parts of the genome where the DNA double strand has separated like a zipper on a jacket to form a sort of loop structure. “We call these R-loops because there’s a third piece of RNA in between the DNA strands.”
The young researcher has been studying R-loops for almost ten years. “They’ve been my passion ever since I wrote my thesis,” she says. As a Sir Henry Wellcome Fellow, she set up the groundwork and carried out experimental research at the MDC before moving to the University of Edinburgh. This culminated in an article in the renowned scientific journal Molecular Cell. “I spent a year and a half in Ana’s laboratory, which was a wonderful time,” says the researcher.
Epigenetic restraint
Skourti-Stathaki explains that not all genes are equally important for the development of a fertilized oocyte into a body cell. Just a few determine the fate of each cell – i.e., which tissue or organ it will transform into. “These genes are critical control programs in all animals and plants,” says the scientist. The developmental genes are controlled epigenetically, which means they are switched on and off indirectly. Nothing changes in the genetic material itself. Instead, enzymes modify the protein scaffold on which the DNA is wound up like a thread.
The helical DNA molecule (light blue) is wound around scaffolding proteins (light yellow and green) like a thread. Together, they form a series of nucleosomes. Polycomb proteins like the Polycomb Repressive Complex (PRC1, dark red, blue, and yellow) bind to this structure and modify the protein portion of the nucleosome chemically.
Skourti-Stathaki suspected that R-loops might play a role in these genes and chose to spend her fellowship working with epigenetics expert Ana Pombo. “Back then, in 2012, I was still in London with my lab,” says Pombo. “Konstantina wrote me an email, and then we met.” The two scientists have been in close contact ever since. “The chemistry between us was right from the start.”
Mysterious proteins
Pombo moved from London to Berlin one year later to join the MDC’s Berlin Institute for Medical Systems Biology (BIMSB). Among other things, she is currently investigating a family of proteins called the Polycomb group, which controls developmental genes epigenetically. Polycomb group proteins switch off these genes, but in such a manner that they are available again very quickly if required. As Pombo puts it: “The cell is only waiting for a trigger to start reading these genes.”
“The need for research in this field is huge, especially in humans and mammals,” says Pombo. “The Polycomb mechanism plays a role in cancer and even in neurological diseases such as schizophrenia.” Skourti-Stathaki sees an important parallel to her research on R-loops. Like Polycomb proteins, R-loops seem to have special, albeit largely unknown, functions in neuronal cells. Skourti-Stathaki explains: “Where R-loops – or the proteins that dissolve them – are missing, severe neurological diseases such as ataxias, a loss of motor coordination, occur”.
New frontiers
The young researcher arrived at the MDC as a postdoctoral fellow in 2015 to start her new project – three years after she first spoke to Ana Pombo. “We wanted to know if R-loops repress certain genes.” says Skourti-Stathaki. “And if these genes are also repressed by Polycomb at the same time, what happens then?”
In order to answer these questions, Skourti-Stathaki sequenced those genome segments that had R-loops or were occupied with Polycomb proteins. “Essentially, nobody was looking into Polycomb and R-loops simultaneously,” she says. Following up on previous work by Pombo and others, she used a system in which development is still in full swing: embryonic stem cells from mice. “We were the first to systematically investigate the function of R-loops and their connection with the Polycomb mechanism in undifferentiated cells,” says Skourti-Stathaki.
Paradoxical results
What they discovered was a kind of security mechanism for developmental genes. A gene repressed by Polycomb and containing an R-loop remains inactivated, they found, even in cases where parts of the Polycomb machinery fail to function. Pombo compares Polycomb proteins to a foot poised on the gas pedal, just waiting to take off. “R-loops are like a brick put under the gas pedal for safety,” she says. In genes untouched by Polycomb, however, the effect is exactly the opposite. In these genes, the R-loops activate the genes. “This time, the brick is on the gas pedal.”
The modified model of Polycomb regulation: R-loops (in the middle) influence the poised transcription machinery (RNA Pol II) as well as Polycomb proteins (PRC1)
“We’ve known about Polycomb for decades,” says Skourti-Stathaki. “And yet this wonderful group of proteins is always full of unexpected surprises.” Until now, the assumed mode of action of these proteins has been fairly simple. The mechanism followed was a fixed sequence of when which individual component of the Polycomb binds to the genome. But the new data question this old model “It’s much more complex,” says Pombo. “There appear to be different levels of repression by the Polycomb proteins.” And R-loops seem to play an important role in this.
It’s the collaboration between two specialists that has made this work so successful
Tasks ahead
But much is still unknown. For example, whether the combination of R-loops and Polycomb is relevant for cancer or neuronal diseases. “Do R-loops make the cell more robust or weaker? We just don’t know,” says Pombo. “This field of research is so young – it is very, very exciting.”
Ana Pombo has to get to her next appointment. She says goodbye to her colleague, puts her cell phone back into her pocket, and closes the lid of her laptop. “It’s the collaboration between two specialists that has made this work so successful,” she says. “Without Konstantina I wouldn’t even have had the idea of looking so closely at this particular area.” She stacks notes and coffee cup back on the laptop, balances them with her left hand, and closes the door to her glass office with her right. Her guest, one of the leading experts in his field, is already waiting for her.
Further information
- Website of the Adrian Bird lab, where Konstantina Skourti-Stathaki is working at the moment
Reference
Konstantina Skourti-Stathaki et al. (2019): “R-Loops Enhance Polycomb Repression at a Subset of Developmental Regulator Genes.” Molecular Cell 73. doi:10.1016/j.molcel.2018.12.016