UPS network
A protein-protein interaction network for the ubiquitin-proteasome system
The ubiquitin-proteasome system (UPS) targets numerous cellular proteins for degradation. In addition, modification of proteins by ubiquitin-like proteins regulates fundamental cellular processes such as cell cycle, DNA repair or gene transcription. Moreover, aberrations in the system have been implicated in the pathogenesis of both inherited and acquired neurodegenerative diseases. Recent studies indicate that the system is involved in the pathogenesis of Parkinson’s, Alzheimer’s and Huntington’s disease. This raises hope for a better understanding of pathogenic mechanisms and the development of novel therapeutic strategies.
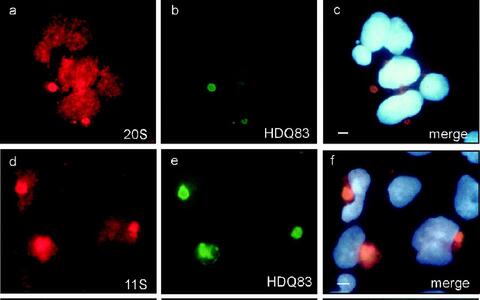
We have demonstrated previously that ubiquitin and parts of the proteasome system are associated with insoluble huntingtin protein aggregates in mammalian cells (Fig. 1). However, the molecular basis for these observations is still largely unknown.
Colocalization of huntingtin protein aggregates with components of the ubiquitin proteasome system (Waelter et al., Mol Biol Cell. 2001 May; 12(5): 1393-1407))
The aim of this project is to connect huntingtin (htt) and its known interacting partners with proteins of the UPS using a high-throughput, automated yeast two-hybrid (Y2H) screening technology. Using this approach we will create a comprehensive protein-protein interaction network for the degeneration of mutant htt in Huntington’s disease. The identified interactions will be validated with systematic luminescence-based assays, co-immunoprecipitations and functional ubiquitination/ deubiquitination assays.
In addition, data will be analyzed using bioinformatic tools and high confidence interaction networks will be created for further experimental validation. The interaction data will be also integrated with expression profiling and proteomics data, along with literature information. Using this strategy, potential functional modules (protein complexes) that are critical for ubiquitination and degradation mutant htt shall be identified.