Selbach Lab
Proteome Dynamics
Profile
Proteins are the central players in this process: First, they are the final product of most genes. Second, the function and dysfunction of proteins is directly responsible for phenotypes. Mass spectrometry-based proteomics is an extremely powerful and versatile approach to study proteome dynamics in biological systems. We are using quantitative mass spectrometry to study the dynamic proteome in health and disease.
Selected research examples
- We globally quantified mammalian gene expression control (Schwanhausser et al., Nature, 2011)
- We analysed the kinetics of cellular protein degradation (McShane et al., Cell, 2016)
- We revealed how mutations in disordered regions affect protein-protein interactions and cause disease (Meyer et al., Cell, 2018)
- We analysed the dynamic proteome of influenza and SARS-CoV-2 infection (Bogdanow et al., Nature Communications, 2019; Wendisch et al., Cell, 2021)
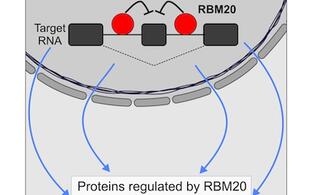
- We studied how phosphorylation of RNA-binding proteins affects their interaction with RNA (Viera-Viera et al., Mol. Cell, 2022)


Team
Research
- 1. From genes to proteins
We are investigating the principles of gene expression control that lead to a specific proteome. Here, we are specifically interested in studying protein synthesis and degradation and the relationship between mRNA and protein levels.
Selected examples
- We globally quantified mammalian gene expression control (Schwanhausser et al., Nature, 2011)
- We analysed the kinetics of cellular protein degradation (McShane et al., Cell, 2016)
- We analysed the dynamic proteome of influenza and SARS-CoV-2 infection (Bogdanow et al., Nature Communications, 2019; Wendisch et al., Cell, 2021)
- 2. From proteins to phenotypes
To assess how the function and dysfunction of proteins affects phenotypes we are studying protein-protein interactions and post translational modifications.
Selected examples
- We revealed how mutations in disordered regions affect protein-protein interactions and cause disease (Meyer et al., Cell, 2018)
- We studied how phosphorylation of ribosomal proteins affects protein translation (Imami et al., 2018)
- We studied how phosphorylation of RNA-binding proteins affects their interaction with RNA (Viera-Viera et al., Mol. Cell, 2022)
- 3. Method development
As a technology-driven lab, we are also developing new methods to facilitate proteome analysis. Frequently, investigating key biomedical questions requires the development of new methods. Therefore, method development is an integral part of most projects, and most of the examples outlined above involve method development. In addition, we are also developing methods that are generally useful for the community
Selected examples
- We developed a new tool that facilitates the design of targeted proteomic methods (Zauber et al, Nature Methods, 2018)
- We present a computational tool that predicts kinase activity in phosphoproteomic datasets (Mari et al., Journal of Proteome Research, 2022)
- We designed a new method to identify drug targets (Melder et al., ChemMedChem, 2022).