Systems Biology of Neural Tissue Differentiation
Profile
With the advent of modern ‘-omics’ approaches, it is becoming increasingly possible to investigate embryonic development systematically and on a genome-wide scale. No longer limited to asking how individual genes are regulated, we can now ask how the genome is regulated to coordinate development. Furthermore, new methodologies allow us to increase the resolution of these approaches – we are now able to resolve how this regulation differs between tissues and even individual cells.
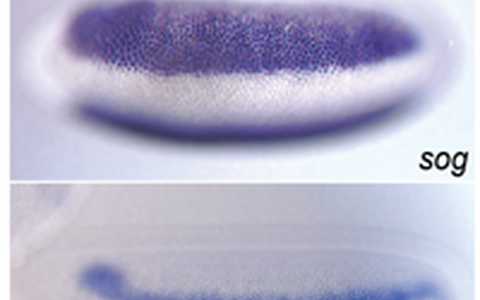
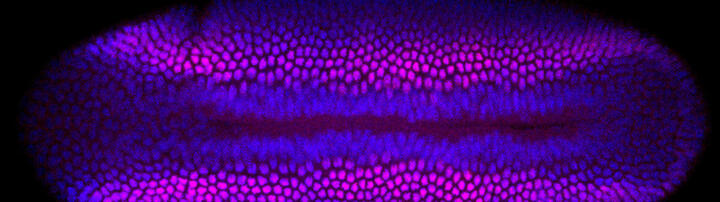
Because of its genetic tractability and experimental accessibility, we approach this question in the context of early embryonic development in the fruit fly Drosophila melanogaster. Using nervous system (NS) development as a model, we focus on how a population of cells in the embryo subdivides into columnar domains that give rise to specific neuroblasts (NBs) and the components of the NS. Our aim is to understand the molecular mechanisms underlying NS primordium specification, its subdivision and the regulated delamination of differentiating NBs.
Research
The cells that make up a complex organism like you can be traced back to a single progenitor – the fertilized egg.
Drosophila neurogenesis – the earliest steps
Over the course of development, however, cells specify and differentiate in a highly regulated manner, so that a complex organism composed of multiple tissues can arise. While individual cell types, such as neurons, epithelial cells, muscle cells, etc. are clearly different from one another, even what we consider to be coherent "tissues", such as the liver, smooth or striated muscle, or the nervous system consist of many distinct cell types. Individual cells differ in terms of size, shape, biochemical properties, and maybe most fundamentally, in terms of their underlying gene expression program.
Our lab wants too understand how cells become different – how do their gene expression programs change over the course of development, and how does this guide and allow for cellular specification and differentiation. Our model to approach this very fundamental question in biology is nervous system development in the fruit fly Drosophila melanogaster.
More than a hundred years of Drosophila research have given crucial insights into some of the key molecules underlying nervous system development, but recent advances allow for a much more systematic, global look at what drives this development. We are employing a systems biology approach to understand nervous system patterning from a genomic and proteomic point of view – we want to understand how key transcription factors regulate gene expression in the developing nervous system, how these transcription factors interact with and shape the epigenetic landscape, but also how such transcription factors act and interact on their cognate enhancers mechanistically.