N. Rajewsky Lab
Systems Biology of Gene Regulatory Elements
Profile
Research Interests and Approaches
The Rajewsky lab studies how RNA regulates gene expression in health and disease. We are collaborating with various clinicians and have built up human brain organoids as a personalized brain disease system. We are also using model systems such as C. elegans, planaria and mouse (including mouse primary neuron culture).
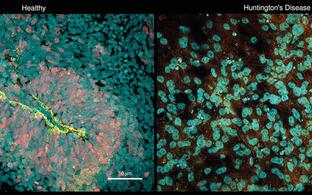
Scientific goals: For understanding the function of RNA in early development, stem cell biology, diseases and regeneration, we traditionally work in a variety of model systems (including C. elegans, Planaria and mice) but in recent years are transforming into medical/clinical systems. For example, we have established patient-derived brain organoids in the lab and study the role of RNA in human brain diseases. We apply single-cell methods and gene-editing or RNA knockdowns by CRISPR directly in brain organoids. We also enjoy direct collaborations with various hospitals from the Charité, such as Neurological Diseases, Pedriatic Cancer, and others.
Methods: We integrate experimental (biochemistry, molecular biology) and computational (bioinformatics, physics, statistics) methods. We have developed several widely used methods (for example, recently, “MirDeep” (Friedlander et al. Nature Biotechnology 2008, Friedlaender et al. NAR 2012); miRNA target identification by chimera analyses (Grosswendt et al. Mol. Cell 2014); fixation method for droplet based single cell sequencing (Alles et al. BMC Biology 2017); “DistMap” for mapping sequenced single cells to correct positions within complex tissues (Karaiskos et al. Science 2017)). A few years ago we implemented and optimized “drop-seq” and are using it in many projects to sequence RNA from single cells in high throughput (e.g. Karaiskos et al. Science 2017, Diag et al. Developmental Cell 2018, Plass et al. Science 2018). We also develop methods to detect, quantify, image, and knockdown/knock out circRNAs (Memczak et al. Nature 2013, Rybak-Wolf et al. Mol. Cell 2014, Piwecka et al. Science 2017). Very recently, we published how to computationally reconstruct cellular spatial positions in tissues from dissociated, single-cell sequenced cells (Nitzan & Karaiskos et al., Nature 2019).
Jobs
We are always looking for skilled people!
Whether your weapon of choice is pipette or keyboard (or both): if you have the impression that this lab is doing interesting research that you would like to contribute to, do not hesitate to contact us.
We are an international and interdisciplinary team (biologists, biochemists, bioinformaticians, physicists, ...) and communication is key. If you are able to (and enjoy) discussing your work with people from other disciplines this lab may be just right!
How to get to our lab
House 101: MDC Berlin-Mitte (BIMSB)
Location: The Rajewsky lab is located in a modern, interdisciplinary and highly collaborative environment: the Berlin Institute for Medical Systems Biology (BIMSB), a new MDC center in the heart of Berlin on the Campus of the Humboldt University, a few hundred meters from the Charité main campus.
Our address
Hannoversche Str. 28, 10115 Berlin
Phone +49 30 9406 2999 (Alex Tschernycheff, Secretary)
Fax +49 30 9406 3868
We are located within a few minutes walk from several public transport stations, such as Oranienburger Tor, Oranienburger Straße, and only one stop away from the public transport hub Friedrichstraße. From Oranienburger Straße, the train S2 connects us directly to the MDC Campus in Berlin-Buch.
S-Bahn: S2 (to MDC Berlin-Buch) and S1, S25, S26
U-Bahn: U6
Tram: M1, M5, 12
Bus: 142
Team
Lab Retreat to MeckPom 2019
As in previous years the group went to the idyllic Gutshaus Rensow in Mecklenburg-Vorpommern in October to enjoy stimulating scientific discussions in a beautiful setting.
Prof. Dr. Nikolaus Rajewsky
Group Leader
Contact information:
- E-mail: rajewsky@mdc-berlin.de
Alex Tschernycheff
Office of the Rajewsky Lab
Contact information:
- E-mail: tschernycheff@mdc-berlin.de
- Phone: +49 30 9406 2999
- Fax: +49 30 9406 3068
Margareta Herzog
Lab Manager
Contact information:
- E-mail: margareta.herzog@mdc-berlin.de
- Phone: +49 30 9406 2996
Denise Aigner
Ph.D. Student
Contact information:
- E-mail: denise.aigner@mdc-berlin.de
- Phone: +49 30 9406 1526
Salah Ayoub
Technician
Contact information:
E-mail: salah.ayoub@mdc-berlin.de
Phone: +49 30 9406 1349
"I am mainly involved in Planaria projects, implementing diverse techniques (RNAi, injections, FACS, etc.). My tasks include lab organization; monitoring and establishment of lab processes, implementation and optimization of protocols, data generation and verification, import/export of biological materials, purchasing of supplies, contact to commercial representatives, management of equipment, next-generation sequencing (NGS) technologies, maintenance of animals (S. mediterranea, C. elegans), and running internal databases (development and administration)."
Anastasiya Boltengagen
Technical Assistant
Contact information:
- E-mail: anastasiya.boltengagen@mdc-berlin.de
- Phone: +49 30 9406 2972
Asija Diag
Postdoctoral Researcher
Contact information:
- E-mail: asija.diag@mdc-berlin.de
- Phone: +49 30 9406 2972
“I’m interested in the spatiotemporal RNA architecture of the germline and how it shapes the proliferation and differentiation balance within this tissue. For this, I use the C. elegans germline as a model system and establish/optimize methods that enable to detect spatiotemporal RNA expression at nearly single cell level.”
Sebastian Ehrig
Postdoctoral Researcher
Contact information:
- Email: sebastian.ehrig@mdc-berlin.de
- Phone: +49 30 9406 1329
"My research aims to investigate how tissue mechanics and local gene-expression pattern influence developmental processes during organoid morphogenesis using combined computational and experimental approaches."
Lisa Emmenegger
Ph.D. Student
Contact information:
- Email: lisa.emmenegger@mdc-berlin.de
"I am interested in studying gene expression regulation in space and time, in the context of early brain development. To do this, I aim to apply CRISPR based approaches, optogenetics and spatial transcriptomics in human brain organoids."
Jonathan Fröhlich
Ph.D. Student
Contact information:
- E-mail: jonathan.froehlich@mdc-berlin.de
- Phone: +49 30 9406 1523
"What phenotypic consequences have mutations in gene regulatory sequences? I have developed an approach for deep mutagenesis of regulatory sequences in the model organism C. elegans. I create hundreds of different mutations and look for their effects on phenotype, fitness and mRNA expression. Our approach will be useful to understand how gene regulatory sequences work in animals."
Talé Heßler
Technical Assistant
Contact information:
- E-mail: tale.hessler@mdc-berlin.de
- Phone: +49 30 9406 2972
Cledi Cerda Jara
Ph.D. Student
Contact information:
- E-mail: cledi.cerdajara@mdc-berlin.de
- Phone: +49 30 9406 2972
"Studying the function of circRNAs in neurons, and the mechanisms underlying their role"
Marvin Jens
Postdoctoral Researcher
Contact information:
- E-mail: marvin.jens@mdc-berlin.de
"I employ and build computational tools to study RNA functionality in space and time, unveil inner states and connections between cells as they form organs and interact with each other in human disease."
Nikolaos Karaiskos
Postdoctoral Researcher
Contact information:
- E-mail: nikolaos.karaiskos@mdc-berlin.de
- Phone: +49 30 9406 1327
"My research focus is the biology at the single-cell level and in particular the study of cell-cell interactions, by using computational and mathematical methods."
Seung Joon Kim
Ph.D. Student
Contact information:
- E-mail: seungjoon.kim@mdc-berlin.de
"I am interested in analyzing single cell RNA-seq data to identify mechanisms involved in pathology of neurological disorders by computational approaches"
David Koppstein
Postdoctoral Researcher
Contact information:
- E-mail: david.koppstein@mdc-berlin.de
- Phone: +49 30 9406 2991
"Alternative RNA isoforms in neurodegenerative disease: The nervous system exhibits a striking degree of alternative splicing and polyadenylation compared to other tissues. These processes are often perturbed in neurodegenerative diseases such as amyotrophic lateral sclerosis and spinal muscular atrophy. I aim to use novel approaches such as long-read, spatial, and single-cell RNA sequencing to determine the extent and dynamics of RNA metabolism dysregulation in such diseases."
Ivano Legnini
Postdoctoral Reseacher
Contact information:
- E-mail: ivano.legnini@mdc-berlin.de
- Phone: +49 30 9406 1524
"I am currently studying the regulation of mRNA metabolism from synthesis to decay, with a particular focus on how different cellular processes such as splicing, export and deadenylation are functionally connected.
On the other hand, I am interested in how genes enforce the spatial organization of tissues, and I am trying to establish innovative methods for manipulating gene expression in space."
Hao Li
Postdoctoral Researcher
Contact information:
- E-mail: hao.li@mdc-berlin.de
- Phone: +49 30 9406 2973
"I am applying spatial transcriptomics and single-cell technologies to understand the mechanism of complex diseases, in particular cancer."
Haiyue Liu
Ph.D. Student
Contact information:
- E-mail: haiyue.liu@mdc-berlin.de
- Phone: +49 30 9406 2989
"My project is on modelling the mRNA kinetics in single cells. We use RNA metabolic labelling method and scRNA-seq technique to capture both new and total mRNA molecules in single cells. With a modified mRNA kinetics model, we fit the mRNA production rate, splicing rate and degradation rate for each gene in single cells across the continuous cell cycle."
Tancredi Massimo Pentimalli
Ph.D. Student
Contact information:
- E-mail: TancrediMassimo.Pentimalli@mdc-berlin.de
"I am interested in understanding the molecular mechanisms underlying tumor initiation, progression/regression and metastasis in a range of solid tumors, including neuroblastoma and breast cancer. In order to identify therapeutic targets and clinical biomarkers, I enjoy merging my medical background with computational data analysis to leverage single cell RNA sequencing and spatial transcriptomic datasets generated from both patient samples and mouse models."
Lena Marie Schott
Technical Assistant
Enes Senel
Ph.D. Student
Contact information:
- E-mail: enes.senel@mdc-berlin.de
"I am interested in using computational techniques and mathematical modelling to explore spatial context of gene regulation and cellular interactions in complex tissues."
Tamas Sztanka-Toth
Ph.D. Student
Contact information:
- E-mail: tamasryszard.sztanka-toth@mdc-berlin.de
- Phone: +49 30 9406 2991
Sarah Samut Tagliaferro
Research Assistant
Contact information:
- E-mail: sarah.samuttagliaferro@mdc-berlin.de, sarah.samut-tagliaferro@bih-charite.de
- Phone: +49 30 9406 1348
"I work in single cell technologies, with particular focus on method development and optimisation in Spatial Transcriptomics."
Ilan Theurillat
Postdoctoral Researcher
Contact information:
- E-mail: Ilan.Theurillat@mdc-berlin.de
- Phone: +49 30 9406 1526
Gwendolin Thomas
Technical Assistant
Contact information:
- E-mail: gwendolin.thomas@mdc-berlin.de
- Phone: +49 30 9406 2972
Marco Uhrig
Ph.D. Student
Contact information:
- E-mail: marco.uhrig@mdc-berlin.de
- Phone: +49 30 9406 1522
"My PhD project is dealing with coding and regulatory functions of small open reading frames (ORFs) that are evolutionary conserved in animals. In order to elucidate their uncharacterized properties and the functional role of encoded micropeptides, I am using a combination of computational and experimental approaches including genome engineering via CRISPR/Cas9 in both cell lines and model organisms, single-cell technologies and human iPS cell-derived brain organoids."
Miriam Wandres
Ph.D. Student
Contact information:
- E-mail: miriam.wandres@mdc-berlin.de
- Phone: +49 30 9406 1325
"I am interested in identifying the underlying molecular mechanisms of diseases in humans. In order to study and understand these mechanisms, I am using human induced pluripotent stem cell derived brain organoids."
Former Lab Members
Agnieszka Rybak-Wolf
was a Postdoctoral Researcher, now Head of Organoid Platform at the BIMSB/MDC, Berlin.
Jonathan Alles
was a Ph.D. Student.
Petar Glazar
was a Postdoctoral Researcher, now working at MPI for Molecular Genetics in Dahlem, Berlin.
Aristotelis Misios
was a Ph.D. Student.
Samantha Praktiknjo
was a Postdoctoral Researcher, now working at BIH.
Marcel Schilling
was a Ph.D. Student.
Janis Hötzel
was a Ph.D. Student.
Monika Piwecka
was a Postdoctoral Researcher, now working as Group Leader at the IBCH PAS in Poznan, Poland.
Mireya Plass Pórtulas
was a Postdoctoral Researcher, now working as Group Leader at the CMRB in Barcelona, Spain.
Christin Sünkel (née Stottmeister)
was a Ph.D. Student.
Kathrin Theil
was a Ph.D. Student.
Panagiotis Papavasileiou
was a Ph.D. Student.
Christine Kocks
was a Senior Scientist and now working with Klaus Rajewsky at the MDC in Berlin-Buch, Germany.
Vera Zywitza
was a Ph.D. Student and now working as Postdoctoral Researcher with Sebastian Diecke at the MDC in Berlin-Buch, Germany.
Sebastian Memczak
was a Ph.D. Student and Postdoc, now working as Postdoctoral Researcher at the Salk Institute in California, USA.
Benedikt Obermayer
was a Postdoctoral Researcher, now working at the BIH in Berlin, Germany.
Andrei Filipchyk
was a Ph.D. Student, now working as Scientific Researcher at the Forschungszentrum Juelich, Germany.
Marta Rodriguez-Orejuela
was a Ph.D. Student.
Luisa Schreyer
was a Technical Assistant.
Jordi Solana Garcia
was a Postdoctoral Researcher, now working as Group Leader at Oxford Brooks University in Oxford, UK.
Filippos Klironomos
was a Postdoctoral Researcher, now working as Senior Scientist at Charité - University Medicine in Berlin, Germany.
Catherine Adamidi
was a Senior Scientist.
Kevin Chen
was a Postdoctoral Researcher, now working as Assistant Professor at Rutgers University, New York City, USA.
Antigoni Elefsinioti
was a Postdoctoral Researcher, now working as Bioinformatics Scientist at Bayer Health Care in Berlin, Germany.
Marc Friedländer
was a Ph.D. Student and Postdoctoral Researcher, now working as Assistant Professor in RNA Biology at Stockholm University, Sweden.
Stefanie Grosswendt
was a Ph.D. Student, now working as Postdoctoral Researcher at the MPI for Genetics in Berlin, Germany.
Dominic Grün
was a Postdoctoral Researcher, now working as Independent Group Leader at the MPI of Immunobiology and Epigenetics in Freiburg, Germany.
Andranik Ivanov
was a Ph.D. Student, now working as Postdoctoral Researcher at the Bioinformatics Core Unit at the BIH in Berlin, Germany.
Anna-Carina Rajewsky (née Jungkamp)
was a Ph.D. Student in the lab, now working for the Federal Ministry of Education and Research in the areas Innovation Policy Issues and Digital Health in Berlin, Germany.
Toshiaki Kogame
was a Ph.D. Student, now working at Kyoto Medical University, Japan.
Azra Krek
was a Ph.D. Student, now working as Senior Research Scientist at Memorial Sloan-Kettering Cancer Center, New York City, USA.
Svetlana Lebedeva
was a Ph.D. Student, now working in the lab of Uwe Ohler at BIMSB/MDC in Berlin, Germany.
Jonas Maaskola
was a Ph.D. Student, now working as Postdoctoral Researcher at KTH Royal Institute of Technology in Stockholm, Sweden.
Sebastian Mackowiak
was a Ph.D. Student, now working as Postdoctoral Researcher at MPI for Molecular Genetics in Berlin, Germany.
Pinar Önal
was a Ph.D. Student, now working as Postdoctoral Researcher with Steve Small at NYU, New York City, USA.
Lena von Oertzen
was a Technical Assistant, now working at the FMP in Berlin-Buch, Germany.
Marlon Stoeckius
was a Ph.D. Student and Postdoctoral Researcher, now working as Co Director R&D at 10x Genomics in Stockholm, Sweden.
Nadine Thierfelder
was a Ph.D. Student, now working as Senior Medical Advisor at Edelman.ergo GmbH in Frankfurt a.M., Germany.
Francesca Torti
was a Ph.D. Student, now working as scientific writer, translator and editor for upwork and GLS Sprachzentrum in Berlin, Germany.
Ulrike Ziebold
was a Postdoctoral Researcher.
Minnie Zhou Fang
was a Postdoctoral Researcher, now working as Executive Director at Wuxi Diagnostics in Shanghai, China.
Research
Examples of research projects
Example 1: Gene expression in the fly embryo at single cell resolution
The lab has a long standing interest in the role of RNA in developmental biology. We established and optimized “drop-seq” in our lab (Alles et al. 2017). This microfluidics based method allows to rapidly capture mRNAs within thousands of individual cells in an hour on a desktop setup. After amplification and sequencing, based on barcode information individual reads can be assigned to individual molecules in individual cells.
In collaboration with the Robert Zinzen lab, we applied this method to the dissociated fly embryo at stage 6. Each of the 6,000 cells of this embryo is distinct as it will give rise to a different part of the fly body. Therefore, it is important to know the molecular makeup of each individual cell.
However, after sequencing individual cells, the problem is to identify the spatial position of this cell within the embryo. We designed a new method that solves this problem (“Distmap”) (Karaiskos et al., Science 2017).
As a result, we quantified gene expression for the vast majority of the embryo cells at a resolution of ~8,000 genes per cell. These data allowed us to create a “virtual embryo” where researchers can perform, online, virtual in situ hybridization for thousands of genes (www.devtex.org).
We identified dozens of transcription factors and lncRNAs as likely novel regulatory factors in this system, and discovered that Hippo signaling is active in the embryo and probably responsible for inducing the exit of patches of cells from synchronized cell divisions, thus explaining observations that have been made decades ago.
Gene expression in the fly embryo at single cell resolution (Karaiskos et al. Science 2017)
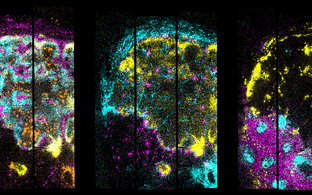
Example 2: Human brain organoids
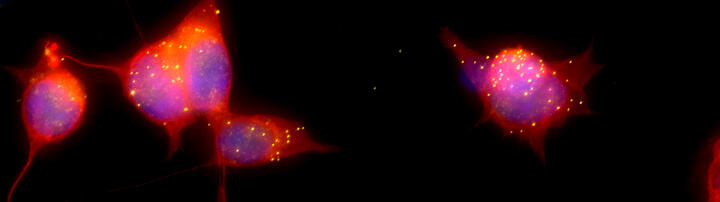
Human brain organoids recapitulate brain development in vitro and enables us to study, at single cell and subcellular level, functions of regulatory RNA in a human-specific genetic background. This allows us to model patient-specific diseases using brain organoids derived from induced pluripotent stem cells (iPSCs) of individual patient. Using the CRISPR-Cas system we can also edit the genome of the pluripotent stem cells in order to introduce or correct disease-related mutations.
In addition, the lab of Nikolaus Rajewsky is working in close collaboration with the Organoid Platform (LINK) on the optimization of protocols for region-specific organoids, such as forebrain organoid protocol (Miriam Wandres, unpublished).
Furthermore, to identify spatial relationships of cells within the 3D brain organoid, clearing and immunolabeling has been recently developed (Anna Ponomarenko and Miriam Wandres, in collaboration with the lab of Andrew Woehler (LINK) and the Organoid Platform, unpublished).
The videos below depict a cleared forebrain organoid labelled for nuclei (grey) and tight junctions (yellow) in both Z-stack and 3D-view - staining and clearing by A. Ponomarenko, organoid generation by M. Wandres and D. Aigner.
Example 3: Regulation by RNA
We are studying regulation by RNA in solid tumors, muscle cells in human patients, and cardio-vascular systems in collaboration with many colleagues.
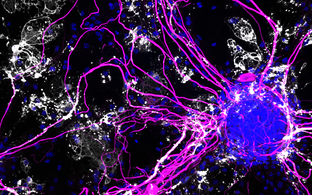
Example 4: Circular RNAs
In 2012/2013, we discovered that animals express a large number of different circular RNAs (circRNAs) in the cytoplasm of cells (Memczak et al. Nature 2013). This observation was also published independently by the Brown and Sharpless labs.
However, we also showed that circRNA expression is tissue and developmental stage specific and identified a circRNA (CDR1as) that we proposed regulates gene expression by binding large numbers of a conserved miRNA (miR-7) (Memczak et al. Nature 2013; back-to-back with the Kjems lab).
Our paper also contained several methods for detecting, imaging, and quantifying circRNA expression. We thus proposed that circRNAs can have regulatory roles. This is interesting since circRNAs are highly stable and therefore may perform specific tasks.
In a serious of follow up papers, we (a) showed that circRNAs are usually produced by “backsplicing” (Ashwal-Fluss et al. Mol Cell 2014) (b) published a widely used public circRNA database where we curate published circRNAs (Glazar et al. RNA 2015) (c) showed that thousands of circRNAs are readily detected in human blood and may serve as biomarkers (d) discovered that a few hundred circRNAs are highly expressed in the mammalian brain and that this expression is well conserved, developmental stage- and tissue specific, and enriched in synaptosomes (Rybak-Wolf et al. Mol. Cell 2015).
Finally, we removed a the CDR1as locus from the mouse genome, showed that the expression of miRNAs that directly interact with CDR1as is specifically altered in the brain of KO mice, and found a behavior phenotype (“pre-pulse inhibition deficit”) that is indicative of neuropsychiatric disorders as well as an electrophysiological synaptic phenotype (Piwecka et al. Science 2017). We believe that these results argue that we discovered a regulatory layer of circRNAs and interacting molecules that have regulatory function in the mammalian brain.
Example 5: Competition effects in post-transcriptional gene regulation
We wrote an “Analysis Review” about functions of competition effects in post-transcriptional gene regulation. In this review, we also proposed and tested one of the first quantitative models. (Jens & Rajewsky, Nature Reviews Genetics 2014).
Model organisms
We study posttranscriptional gene regulation in cell culture and two famous model systems: The nematode Caenorhabditis elegans (and its relative Caenorhabditis briggsae) and Schmidtea mediterranea.
C. elegans
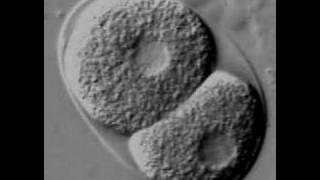
The early embryo and the germline of C.elegans are highly regulated on the posttranscriptional level. For example, while PolII activity starts at the 4-cell stage, the earlier divisions already differentiate the future germline from the soma and set the body axes, without any transcription at all. [Stitzel and Seydoux 2007, Yamamoto, I.; Kosinski, M. E. & Greenstein, D. (2006)]
- Background information
-
An ideal system to explore very early developmental processes
-
Since its establishment as a model organism, Caenorhabditis elegans has been an invaluable tool for biological research. An immense spectrum of questions can be addressed using this small nematode, making it one of the most versatile and exciting model organisms.The worm is also the organism in which miRNAs have been first described.
The worm is an ideal system to explore the very early developmental processes because of its rapid and invariant development and its ease of RNAi and transgenic techniques. Studies in the model organism C. elegans have yielded most of what is known today about metazoan development. The first embryonic division in C. elegans has been intensely studied since it is used as an in vivo system for detailed studies of metazoan cell division.
Very early development in the worm (and most other species) is mainly controlled by maternal gene products which are loaded into the oocyte during oogenesis. After fertilization the C. elegans embryo undergoes a series of stereotyped asymmetric cleavages that spatially segregate these maternal factors, such as transcription factors and coding and noncoding RNAs. After a set of divisions the embryo activates its own transcription. This so called maternal to zygotic transition is a universal process in animal development: The embryo overtakes the control and thus no longer solely relies on maternally provided transcripts.
We are investigating the function of small RNAs during very early development of C. elegans.
S. mediterranea
The fresh water flatworm S. mediterranea has been described already more than 200 years ago. Like many flatworms, it exhibits an astounding capacity to regenerate after injuries: a single animal, cut into dozens of pieces, may give rise to dozens of fully regenerated, clonal animals.
Also, unlike another famous regeneration model organism, the radial-symmetric Hydra, the bilateral planarians are complex, with an organized nervous system, eyes and other organs.
The young Charles Darwin collected some land planarians during his voyage with the HMS Beagle (1831 to 1836). He notes:
"Having cut one of them transversely into two nearly equal parts, in the course of a fortnight both had the shape of perfect animals. I had, however, so divided the body, that one of the halves contained both the inferior orifices, and the other, in consequence, none. In the course of twenty-five days from the operation, the more perfect half could not have been distinguished from any other specimen.
The other had increased much in size; and towards its posterior end, a clear space was formed in the parenchymatous mass, in which a rudimentary cup-shaped mouth could clearly be distinguished; on the under surface, however, no corresponding slit was yet open. If the increased heat of the weather, as we approached the equator, had not destroyed all the individuals, there can be no doubt that this last step would have completed in structure. Although so well known an experiment, it was interesting to watch the gradual production of every essential organ, out of the simple extremity of another animal."
["The Voyage of the Beagle" pp.26]

- Background information
-
-
The Rajewsky lab established planaria as a model system in the lab. These freshwater flatworms are famous for their almost unlimited ability to regenerate any tissue via pluripotent, adult stem cells. We are studying the role of small RNAs in planarian regeneration.
Publications
News
Data, Software & Resources
- Discrover
-
A motif discovery method to find binding sites of nucleic acid binding proteins
-
Find further details on the Discrover website.
The corresponding publication is:
"Binding site discovery from nucleic acid sequences by discriminative learning of hidden Markov models"
Jonas Maaskola and Nikolaus Rajewsky
Nucleic Acid Research, 42(21):12995-13011, Dec 2014. doi:10.1093/nar/gku1083 - Planaria
-
Useful resources for the planarian research community
-
Gene models
The transcripts sequences were obtained by genome independent de novo transcriptome assembly (Adamidi et al.(2011)) combined with genome based transcript predictions inferred with CUFFLINKS (Trapnell et al. (2010)). For details, see Adamidi et al. (2011) and Oenal et al. (2012).
Genomic transcript coordinates
If a transcripts maps to multiple genomic contigs, only the longest matching region is reported.
Expression data
Oenal_et_al_2012_mRNA_and_protein_expression_data.csv
The file contains transcript and protein expression data for all genes. If the protein expression was quantified for peptides in different translated frames (due to frame-shifting errors in the genome independent de novo annotation) the table contains multiple entries for the respective gene.
See the README for details on the table format.
- eFACS
-
A method to collect staged C. elegans embryos by fluorescence-activated cell sorting
-
We devised a method to collect staged C. elegans embryos by fluorescence-activated cell sorting (eFACS). A single eFACS run routinely yields tens of thousands of almost perfectly staged one-cell stage embryos.
In principle, eFACS can be used to extract large samples of embryos enriched in any desired embryonic stage. Thus, eFACS opens the door to apply many modern high-throughput technologies to assay embryonic developmental stage specific gene expression in C. elegans. Several of such investigations are already ongoing in the lab.
- iPAR-CLIP
-
A PAR-CLIP protocol for living C.elegans animals
-
Our lab established a PAR-CLIP protocol for living C.elegans animals, called iPAR-CLIP. This allows to study RNA-binding protein interactions in vivo. Here, we list our published work, using this method. All sequencing data of published experiments are available through GEO.
In Vivo and Transcriptome-wide Identification of RNA Binding Protein Target Sites
Anna-Carina Jungkamp , Marlon Stoeckius , Desirea Mecenas , Dominic Grün , Guido Mastrobuoni , Stefan Kempa , Nikolaus Rajewsky
Mol Cell. 2011 Dec 9;44(5):828-40. doi: 10.1016/j.molcel.2011.11.009.
Unambiguous identification of miRNA:target site interactions by different types of ligation reactions.
Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N.
Mol Cell. 2014 Jun 19;54(6):1042-54. doi: 10.1016/j.molcel.2014.03.049. Epub 2014 May 22.
A variety of dicer substrates in human and C. elegans.
Rybak-Wolf A, Jens M, Murakawa Y, Herzog M, Landthaler M, Rajewsky N
Cell. 2014 Nov 20;159(5):1153-67. doi: 10.1016/j.cell.2014.10.040.
A note on RNA thio labeling
The above papers utilized 4-thiouridine for RNA-labeling and crosslinking. We have also attempted to use 4-thiouracil as an alternative to 4-thiouridine for iPAR-CLIP in C.elegans. The raw data can be made available upon request by E-mail to Nikolaus Rajewsky.
- PicTar
-
An algorithm for the identification of microRNA targets
-
PicTar is a project of the Rajewsky lab at NYU's Center for Comparative Functional Genomics and the MDC, Berlin.
The PicTar website provides details (3' UTR alignments with predicted sites, links to various public databases etc) regarding:
- microRNA target predictions in vertebrates (Krek et al, Nature Genetics 37:495-500 (2005))
- microRNA target predictions in seven Drosophila species (Grün et al, PLoS Comp. Biol. 1:e13 (2005))
- microRNA targets in three nematode species (Lall et al, Current Biology 16, 1-12 (2006))
- human microRNA targets that are not conserved but co-expressed (i.e. the microRNA and mRNA are expressed in the same tissue) (Chen and Rajewsky, Nat Genet 38, 1452-1456 (2006))
- UPDATED PICTAR PREDICTIONS 2012: doRiNA: a database of RNA interactions in post-transcriptional regulation (Anders et al, Nucleic Acids Res. 2012 Jan;40(Database issue):D180-6. Epub 2011 Nov 15.)
Bulk Downloads
PicTar miRNA target site predictions for the hg17, mm7, dm2 and ce2 genomes can be obtained from the UCSC genome browser via the 'tables' feature.
New predictions for hg18, mm9 and ce6 are available on our own UCSC mirror at http://dorina.mdc-berlin.deFor your convenience, doRiNA offers a link for downloading PicTar target site predictions: simply run a doRiNA query for all miRNA targets, setting the search options to 100% under step 3, and follow the link on the top of the results page.
AlignmentBuilder
To build contiguous alignments for each 3' UTR or other types of mRNAs (5' UTRs, CDS, ..) from the MultiZ multiple alignments at the UCSC Genome Database you can use the scripts from AlignmentBuilder.
Notes
- The format of the gene annotation table available from UCSC changes occasionally. Check that the strand is given as "+" or "-", and not just "+" or blank. In the latter case you need to insert the missing "-"
- The program also requires that the gene annotation table have exactly 10 fields - no "bin" field at the beginning and no extra fields at the end. Currently, there is a short script, transform_annotation.py that removes the first field and the extra fields at the end, but watch this in the future in case the format changes again.
- There is an off-by-one error with the multiZ alignments file. You need to add 1 to the second field of that file or the alignments will be off by one.
- Do not use the characters "." or "-" in the gene name because this will cause errors.
- miReduce
-
-
miReduce correlates the logarithm of expression fold changes of a set of genes with the motif content of the regulatory sequences of these genes. In particular, it correlates the genome wide mRNA log fold changes in experiments involving the knockout/knockdown/overexpression of one or more miRNAs with the motif content of the 3'UTRs. The model is based on linear regression, and is intended for genome-wide studies.
The code is written as a Perl script for Linux. The details on how to use it and how to interpret the results are in the README file, as well as the literature where more details can be found.
To unpack, do the 'tar -xzf miReduce.tgz'. This will create a directory called miReduce.
If you decide to try and use miReduce, please read the README file carefully, as this is a somewhat rough implementation that we still decided to release due to high demand for this kind of tool.
- RNA competition
-
-
We here provide data and code to reproduce the analyses and plots from the following publication:
“Competition between target sites of regulators shapes post-transcriptional gene regulation”.
Nature Reviews Genetics (Analysis) 2014 (in press, doi:10.1038/nrg3853) Jens M, Rajewsky N.Download
The tar archive contains python scripts that work with python 2.7, numpy, and matplotlib. Please see the contained README and LICENSE for details.
You can download the tarball here.
- miRDeep2
-
Discovering known and novel miRNAs from deep sequencing data
-
Developed by Sebastian Mackowiak, Marc Friedländer and Nikolaus Rajewsky
Systems Biology group at the Max Delbrück Center, Berlin-Buch
April 20th, 2011miRDeep2 is a completely overhauled tool which discovers microRNA genes by analyzing sequenced RNAs. The tool reports known and hundreds of novel microRNAs with high accuracy in seven species representing the major animal clades. The low consumption of time and memory combined with user-friendly interactive graphic output makes miRDeep2 accessible for straightforward application in current research.
Download the latest release from here: https://github.com/rajewsky-lab/mirdeep2/releases/latest (v0.1.2, updated 27/02/2019)
The package is freely available for non-commercial purposes. For commercial purposes, please contact us.
For general comments or questions on commercial use, contact: rajewsky at mdc-berlin dot de.
Disclaimer
The miRDeep2 software package is provided as is without any guarantees or warranty for correctness. The authors are not responsible for any damage or loss of any kind caused by the use or misuse of the scripts included in the software package. The authors are not under obligation to provide support, service, corrections, or upgrades to the package.
miRDeep
Developed by Marc Friedländer and Nikolaus Rajewsky
Systems Biology group at the Max Delbrück Center, Berlin-Buch
April 4th, 2008The miRDeep package was developed to discover active known or novel miRNAs from deep sequencing data (Solexa/Illumina, 454, ...). The package consists of everything you need to analyze your own deep sequencing data after removal of ligation adapters: a number of scripts to preprocess the mapped data, and the core miRDeep algorithm that will analyze and score these data.
The package is freely available for non-commercial purposes. For commercial purposes, please contact us.
If you use miRDeep in your scientific research, please cite us:
Friedländer, M.R., Chen, W., Adamidi, C., Maaskola, J., Einspanier, R., Knespel, S., Rajewsky, N. 'Discovering microRNAs from deep sequencing data using miRDeep', Nature Biotechnology, 26, 407-415 (2008)
Three compressed downloads are available at this site:
Both demo_limited and demo_full contain a README file that contains instructions how to run miRDeep on your own deep sequencing data. Running it on the demo data allows you to test that it produces what it is supposed to produce (positive control). We highly recommend to test the package on the demo data.
- miRDeep.tgz contains the core algorithm script and all auxiliary scripts.
- demo_limited.tgz contains all the files that we used to predict C. elegans known and novel miRNA, starting with the file containing the alignments of the filtered reads to the nematode genome and ending with the file of the final predictions (size of this demo: a few MB)
- demo_full.tgz contains all the files that we used to predict C. elegans known and novel miRNA, starting with the file containing the alignments of all reads to the nematode genome and ending with the file of the final predictions (warning: size of this file: 100 MB)
For general comments or questions on commercial use, contact: rajewsky at mdc-berlin dot de
(substitute 'at' with '@' and 'dot' with '.' to get the proper email addresses)
Older miRDeep2 versions
Previous releases can be found here: https://github.com/rajewsky-lab/mirdeep2/releases
- pSILAC
-
-
For the pSILAC webpage follow the link to http://:psilac.mdc-berlin.de