Ubx2 links the Cdc48 complex to ER-associated protein degradation
The secretory pathway of eukaryotic cells harbors an elaborate protein quality control system, which prevents the accumulation of misfolded or unassembled proteins in the secretory pathway. This system is localized in the Endoplasmic Reticulum (ER). ER associated protein degradation (ERAD) is an important component of this quality assurance system and directs misfolded proteins for destruction by the cytoplasmic ubiquitin-proteasome pathway. ERAD can be divided mechanistically into separate steps: First, misfolded proteins are detected within the ER-lumen. Second, the proteolytic substrates are targeted to and inserted into an aqueous transport channel. Third, the substrates are transported back into the cytosol in a process termed dislocation or retro-translocation. Fourth, components of the cytosolic ubiquitin system synthesize a polyubiquitin chain on the dislocated substrates. Fifth, the AAA-ATPase complex consisting of Cdc48p, Ufd1p, and Npl4p helps to release the ubiquitin-conjugated substrates from the ER-membrane. Finally, the cytosolic 26S-proteasome digests the ubiquitin-conjugated dislocated molecules.
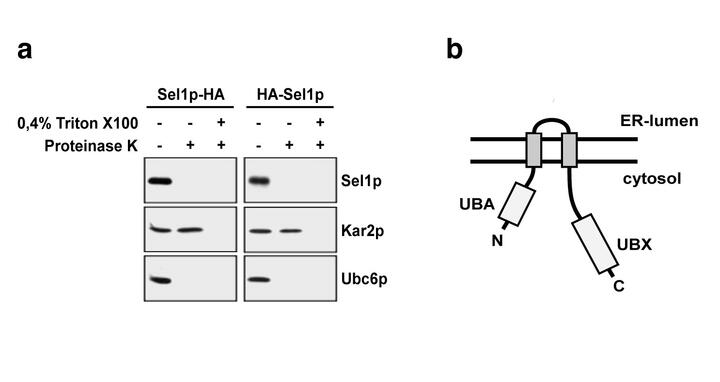
Now, Oliver Neuber (laboratory of Dr. Thomas Sommer) and colleagues have identified a new component necessary for ERAD (Nature Cell Biology, doi: 10.1038/ncb1298). This factor, Ubx2/Sel1p, recruits the assembled Cdc48p-complex to the ER-membrane-bound ubiquitin ligases Doa10p and Hrd1p. Proteolysis of membrane-bound and ER-luminal ERAD substrates is strongly reduced in cells lacking Ubx2/Sel1p, demonstrating that the Ubx2/Sel1p mediated interaction of the Cdc48p/p97 complex with the ubiquitin ligases is crucial for ERAD. This view is further strengthened by the fact that in ubx2/sel1 knockout cells the ER-stress response (unfolded protein response, UPR) is turned on and that UBX2/SEL1 transcription is up-regulated in response to ER-stress. In addition, the scientists have shown that Ubx2/Sel1p function is not limited to ERAD substrates but is also involved in breakdown of cytosolic substrates that are processed by membrane-bound ubiquitin ligases.
Ubx2/Sel1p belongs to the family of UBX-domain proteins, which have been shown to interact directly with Cdc48p. Yeast Ubx2/Sel1p is unique since it comprises not only an UBX domain but an ubiquitin-binding domain (UBA) in addition. Both domains are separated by two transmembrane segments, which anchor Ubx2/Sel1p in the ER-membrane. Based on experimental data, the researchers propose that Ubx2/Sel1p acts upstream of the Cdc48p ATPase complex in the degradation of substrates of the ubiquitin proteasome pathway in general. Furthermore, their data suggest that the UBA-domain of Ubx2/Sel1p may be specifically required for the release of ERAD substrates from the membrane but not for degradation of cytosolic substrates. Ubiquitin-binding domains are common motives of ERAD components. Thus, it is tempting to speculate that ERAD substrates are guided from a dislocation channel to the proteasome by a cascade of ubiquitin-binding factors, including Ubx2/Sel1p. These successive interactions may protect the substrate from premature de-ubiquitination and in addition contribute to the directionality of the transport process.
Contact:
Pamela Cohen
p.cohen@mdc-berlin.de
+49 30 9406 2121