Electron Microscopy
Séverine Kunz
Profile
Our group offers a set of methods to visualize organelle ultrastructure as well as the surrounding context at nanoscale resolution. We explore manifold specimen types spanning from cells and organoids to model organisms, such as mice, zebrafish, and fruit flies. Imaging modalities using Transmission EM (TEM) and Scanning EM (SEM) generate images which result in 2D or 3D data. Additional to analysis of the cellular ultrastructure, immuno-EM techniques or correlative light and electron microscopy (CLEM) methods combine functional information on protein identity with the underlying morphological features at nanometer resolution.
Team
Methods and Applications
Methods
- Sample preparation
The term sample preparation encompasses all procedures a specimen undergoes prior to imaging with electron microscopy. Proper workflows and high-quality preparation are essential to ensure optimal imaging results. Poor sample preparation can significantly distort data interpretation.
Sample preparation techniques:
- Negative staining: the process in which isolated proteins, macromolecular complexes or cell fractions are contrasted with heavy metals
- Fixation: the process of arresting cell activity and preserving ultrastructure or topology
- Chemical aldehyde fixation is suitable for cell monolayers, cell pellets, small organisms, tissues, organoids, biopsies that are 1-2 mm3
- High-pressure freezing ensures vitrification of the sample for better structural preservation and it is suitable for cell monolayers, cell pellets, small organisms, tissues, organoids and biopsies with the maximum thickness of 200µm and 5.5 mm diameter.
- Plunge-freezing ensure vitrification of samples and is suitable for isolated proteins, macromolecular complexes and cell monolayers
- HMDS sample preparation for scanning electron microscopy as a replacement for critical point drying
- Dissection
- Embedding: the process in which samples are contrasted, dehydrated and stabilized mechanically in a resin. We use epoxy and acrylic resins.
- Manual embedding
- Microwave embedding
- Freeze substitution
Sample preparation devices:
- Ted Pella Biowave: microwave
- Leica EM ICE: high-pressure freezing
- Leica AFS: freeze substitution
- Leica FSP: freeze substitution and embedding in acrylic resins
- Leica MZ75: stereomicroscope
- Ultramicrotomy
Ultramicrotomy is the technique used to section resin-embedded samples into ultra-thin slices for electron microscopy. Using diamond knives, specimens are cut and placed onto grids for transmission electron microscopy (TEM) or onto solid substrates for scanning electron microscopy (SEM). Following sectioning, samples are typically post-stained with heavy metals to enhance image contrast.
Microtomes:
- Leica UC7 with cryo-chamber for Tokuyasu sectioning
- Reichert Ultracut S – two devices
- Reichert Ultracut E
Other devices:
- GloQube glow discharge system
- Safematic carbon and sputter coater
- Leica AC20: grid stainer
- Imaging
Our technology platform provides a comprehensive suite of electron microscopy techniques tailored to the needs of the MDC research community. We are able to image a wide range of specimens – from human biopsies to isolated proteins. We support imaging of a wide range of specimens—from human biopsies to isolated proteins—using both transmission electron microscopy (TEM) and scanning electron microscopy (SEM). These modalities enable high-resolution visualization of cellular ultrastructure and organelle interactions. To bridge molecular identity with structural context, we integrate EM with advanced complementary techniques such as immuno-labelling and fluorescence light microscopy.
Transmission Electron Microscopy (TEM)
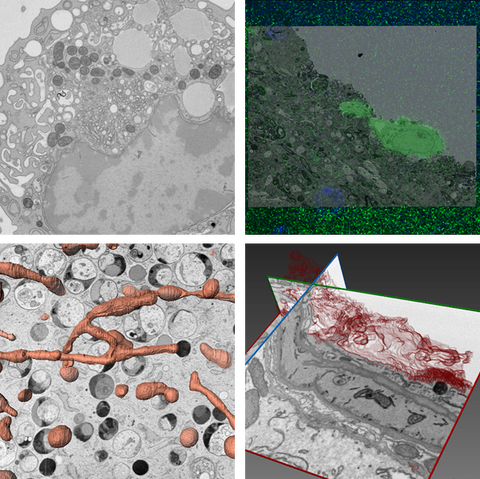
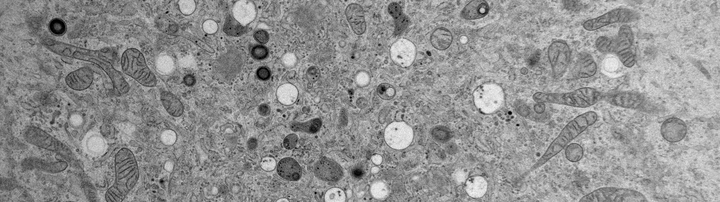
In TEM a beam of electrons is transmitted through a thin sample (50-250 nm). TEM allows for high resolution visualization of cell morphology and ultrastructure. Recorded micrographs are 2D projections of 3D volumes and can be tricky to interpret. For studying complex interactions, such as between cells or organelles, electron tomography (ET) is employed. In ET, the sample is tilted within the microscope (±70°), and a tilt series is recorded. This image stack is then reconstructed into a 3D volume (tomogram), revealing spatial relationships that are obscured in 2D projections.
Scanning Electron Microscopy (SEM)
In scanning electron microscopy, a focused beam of electrons is used to scan the surface of a specimen. Different detectors allow for the collection of back-scattered electrons (BSE) and of secondary electrons (SE). SEM can be used to study ultrastructure by imaging resin sections mounted on a solid substrate or the topology and surface morphology of whole cells.
Imaging modalities
- 2D Imaging:
- TEM of negative stained grids, method used in structural biology for assessing sample quality, typically an intermediate step before moving to cryo-EM
- TEM of resin sections
- SEM of resin sections
- TEM montages using SerialEM
- SEM large scale tile scans using ThermoFisher MAPS software
- 3D Imaging:
- Electron tomography: TEM of sections up to 200-220 nm thick
- Array tomography: SEM imaging of array of sections allowing for volume EM (vEM) of whole cells, small organisms, tissues. Produces anisotropic voxels (5x5x70nm)
- Cross-section milling: FIB-SEM technique suitable for vEM of subcellular structures. Milling can be done with Gallium or with plasma. Produces isotropic voxels (5x5x5 nm)
- Spin milling: large-area planar plasma milling of resin blocks, similar geometry as a serial-block face setup (e.g 3View), but with capability of reaching isotropic voxels (5x5x5 nm)
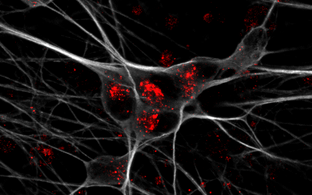
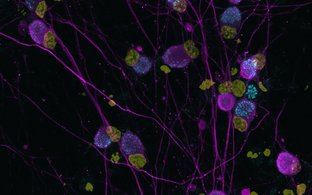
- Correlative light and electron microscopy
- Pre-embedding CLEM: light microscopy data and and region-of-interest mapping performed before EM sample preparation
In-resin CLEM: advanced sample preparation using high-pressure freezing and freeze substitution into an acrylic resin preserves fluorophores. Sections are imaged using light microscopy followed by electron microscopy
Imaging devices
- Transmission electron microscopes:
- Jeol JEM 2100 Plus (200kV), 20MP CMOS camera Xarosa with SerialEM
- ThermoFisher Talos L120 C (120 kV) with long duration dewar for cryo applications, 16M Ceta CMOS camera and 2 Gatan cryo holders (AG Daumke)
- Zeiss Leo 910 (80 kV) with 11M Quemesa CCD camera
- Scanning electron microscopes:
- ThermoFisher Helios 5CX Hydra Dual-beam system with plasma-FIB (xenon, oxygen, argon and nitrogen)
- ThermoFisher Helios 5CX with Elstar column and Tomahawk Gallium-Fib equipped with in-column, in-lens, retractable DBS and STEM detectors. This is a shared investment with the Leibniz Institute for Molecular Pharmacology (FMP)
- 2D Imaging:
- Data analysis light
EM data analysis is crucial because it allows for the transformation of raw, high-resolution micrographs into meaningful scientific insights. Ideally, both imaging and analysis are conducted in a blinded manner by multiple users, with conclusions drawn only after comprehensive data evaluation.
We offer full support for qualitative analysis and limited support for quantitative analysis, including guidance on quantification strategies, workflow setup, and supervision.
- Qualitative analysis: refers to any non-numerical interpretation of micrographs (e.g identification of cell types, organelles, morphological description etc)
- Quantitative morphometric analysis: can be done on 2D and 3D data and implies measuring morphological parameters of structures of interest (e.g area, perimeter, length, volume, brenching etc)
- Stereology: a statistical method that allows extraction of 3D quantitative information from 2D images and is used for calculation volume fractions of organelles (e.g the mitochondrial network represents 38% of total cardiomyocyte volume)
- Correlation: applied in correlative imaging projects to align different modalities (e.g., fluorescence and EM) for precise localization of structures of interest.
- Tomogram reconstruction: generation a 3D volume for an aligned tilt series obtained from tomography
- Object annotation and segmentation: identification and labeling of specific structures (e.g organelles) in 2D micrographs or 3D volumes. Manual annotation is a crucial step for generating the ground truth that will be used for training of a neural network. Advanced annotation and network training can be accessed through Helmholtz Imaging
- Visualization: rendering and displaying segmented structures for interpretation, quantitavice analysis and presentation