Bunina Lab
Systems biology of cardiovascular and neuronal pathologies
Profile
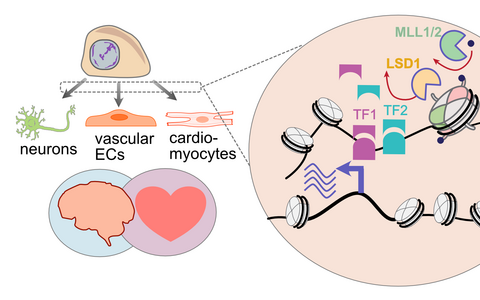
Different cells in our bodies form distinct tissues and organs to perform specific functions, even though each cell carries the same genetic material in its DNA. DNA does not exist alone in the cells but is closely associated with a class of proteins called histones. Together DNA and histones form the physiological form of our genome – chromatin. Histones carry specific protein modifications and their precise positioning on the DNA controls the activity of regulatory regions and expression of genes during cell differentiation, making epigenetic landscape one of the crucial factors in defining cell types and their developmental stages.
Our group studies how malfunctioning epigenetic regulation leads to the pathogenesis of both cardiovascular and neurodevelopmental disorders. In a systemic approach we are combining bulk and single-cell “omics” technologies and novel data integration methods to dissect gene regulatory networks and identify molecular pathways perturbed in human disease. As a model system we use human induced pluripotent stem cells and derived from them differentiated cells (neurons, vascular cells, cardiomyocytes) and 3D cell cultures.
Team
Group retreat 2025
Team photo 2023
Alumni
Jennifer Sanmartin (summer intern) - BSc in computer science, Princeton University, US
Thadoe Thukha (Master student) - now PhD student at Heidelberg University, Germany
Eileen Kuhnke (Bachelor student)
Vanessa Gimmel (Bachelor student)
Elizaveta Kulaeva (Master internship) - now PhD student at Princess Maxima Center, Netherlands
Research
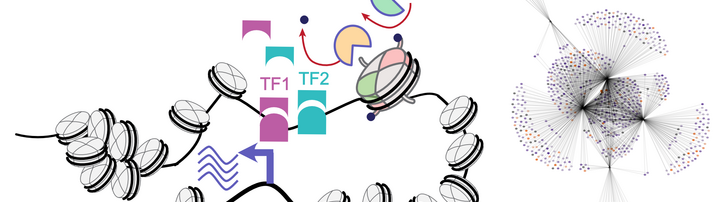
The epigenetic landscape is a crucial factor in defining cell types and their developmental stages, where the precise positioning of chromatin modifications controls the activity of regulatory regions and gene expression during cell differentiation. Mutations in chromatin-modifying proteins, such as histone demethylase LSD1 and methyltransferases MLL1/2, underlie the pathogenesis of neurodevelopmental disorders coupled with vascular malformations. Although cardiovascular and nervous systems are highly interconnected and co-regulated, it is unclear why mutations in chromatin factors affect both systems. Our research focus is on dissecting the interplay between chromatin, transcription factors and their target genes in regulating cell state transitions in health and disease.
The central question of our group's research is how chromatin-associated networks affects cell differentiation and contribute to neurodevelopmental and cardiovascular disorders.
Currently we're pursuing three main research directions:
- Dissecting chromatin network dysfunction in rare monogenic neurodevelopmental disorders with cardiovascular impairement
The malfunctioning of all H3K4 methyltransferases (such as MLL1 and MLL2) and demethylases (LSD1) is frequently found in cancer and, more recently, in rare neurodevelopmental disorders, such as Wiedemann-Steiner syndrome (MLL1 mutations), Kabuki syndrome (MLL2 mutations), and CPRF syndrome (LSD1 mutations). In our recent work we discovered impaired pluripotency network dynamics during early differentiation of patient-derived LSD1 mutated cells (Bunina et al, 2021). In current projects we are using CRISPR-edited isogenic iPSCs and multi-omic profiling during in vitro differentiation of iPSCs to neuronal and cardiovascular cell types, we aim to identify lineage-specific and shared vulnerabilities, misregulated transcription factors, and chromatin state transitions that underlie developmental defects and uncover how cellular microenvironment modulates this misregulation.
- Uncovering the epigenetic basis of endothelial cell heterogeneity across tissues
Vascular endothelial cells show remarkable functional and molecular heterogeneity, largely attributed to microenvironment and paracrine signaling, but little is know about the epigenetic regulators of angiodiversity (Trimm and Red-Horse, 2022). Here by integrating enhancer-driven gene regulatory network inference with machine learning approaches, we aim to identify tissue-specific epigenetic signatures and candidate regulatory elements which we could functionally validate.
- Vascular biomarkers of ischemic stroke response
Ischemic stroke is a cerebrovascular disease, with brain vasculature playing an important role in its progression. Previously shown epigenetic reprogramming in endothelial cells after stroke suggests the important function of the chromatin states in the long-term endothelial recovery due to epigenetic memory (Felling and Song, 2014). Despite tremendous progress in stroke research in the past decades, the lack of functional annotation for enhancer elements in the post-ischemia/reperfusion stage limits biomarker studies for vascular phenotypes, leaving many non-coding genetic variants associated with vascular traits unexplored. This knowledge gap hinders the identification of target molecules as potential biomarkers for post-ischemic prognostics and the development of targeted therapies to mitigate or reverse endothelial dysfunction. In this project we use experimental and computational omics approaches, as well as AI models to extract epigenomic response to metabolic perturbation model of stroke.