Kühn Lab
Genome Engineering & Disease Models
Profile
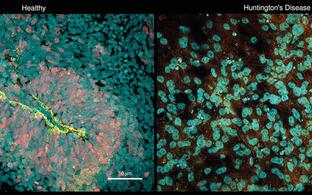
We use advanced CRISPR-Cas9 technologies, in particular our patented REPLACE approach* (Danner et al. 2021) for genome engineering in mammalian cells. Using these tools we build new alleles for disease modeling and repair mutations for therapy in mouse and human model systems.
Specifically we are running projects to:
- build new disease models in mice and human iPS cells
- approach gene therapy in mouse models and patient-derived cells
- explore exon replacement by NHEJ for gene repair and humanisation
- operate the core facility for mouse genome engineering
- pursue genome repair to optimize the C57BL/6 inbred mouse strain
Genome engineering is a key technology to study gene function in health and disease. The continuous innovation of gene editing technologies is the base for the next generation of biomedical research.
*Nucleic Acid Sequence Replacement by NHEJ. PCT International Publication Number: WO 2019/122302 A1 (2019).