How RNA-binding proteins are regulated
When our cells need a specific protein, the corresponding gene is first read and its DNA sequence is transcribed into RNA. “Research has long focused heavily on the required transcription factors, that is, on how the immediate synthesis of RNA is controlled. But this is just the first step of gene expression. What comes after that – post-transcriptional regulation – is also a very important process,” explains Professor Matthias Selbach, head of the Proteome Dynamics Lab at the Berlin-based Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC) and the last author of a paper on RBPs now appearing in the journal “Molecular Cell”.
The next step involves RNA-binding proteins, which are the focus of the study. In translation, which occurs outside the cell nucleus in the surrounding cytoplasm, the RNA – now in the form of messenger RNA – acts as a blueprint used by ribosomes to synthesize proteins from amino acids. RNA-binding proteins make sure that mRNA is transported from the cell nucleus, is guided to the ribosome, stabilizes, and can be broken down again when it is no longer necessary.
In contrast to transcription factors, however, relatively little is known about how the approximately 1,900 RNA-binding proteins are regulated. Most of the time, protein functions are manipulated by chemical modifications – such as through the targeted addition of phosphate groups to individual amino acids (phosphorylation). This usually changes the activity of the protein.
Important for titin synthesis
In recent years, scientists have found more than such 100,000 phosphorylation sites on proteins. “But for the vast majority of them, we don’t know what the phosphate groups do functionally,” Selbach says. So it is high time to take a closer look at RNA-binding proteins. RBM20, which is being intensively studied at the MDC, is an RNA-binding “master regulator.” It especially plays a crucial role in the synthesis of the variant-rich, elastic protein titin in the heart muscle, as well as in the pathogenesis of heart muscle diseases. Dr. Carlos Henrique Vieira e Vieira, lead author of the study, used a method developed by Professor Markus Landthaler at the MDC called RNA interactome capture – or RIC for short – and extended it to incorporate a quantitative analysis method called qRIC.
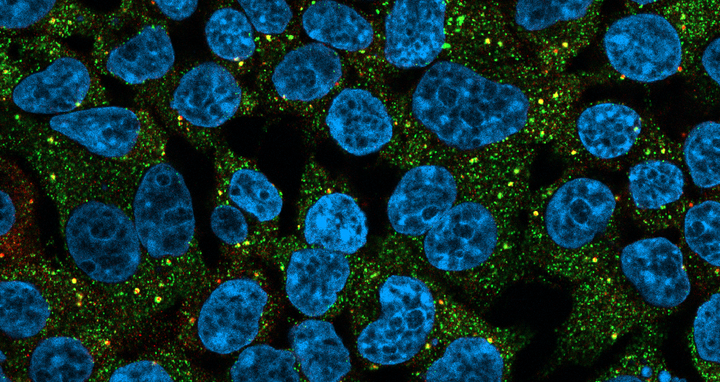
The image shows human cells with a disease-causing RBM20 mutant: the cell nuclei are shown in light blue, the RBM20 protein in green and the MOV10 protein in red. This protein marks granular organelles in the cells that contain RNA, which marks RNA-containing granular organelles in the cells.
Proteins are normally bound together by weak forces, such as electrostatic interactions and hydrogen bonds, when interacting with RNA. “By using UV light we can ‘freeze’ the cell in this state – and only once this is accomplished do we unlock it,” Selbach explains. This is achieved by irradiating living cells with UV light. All proteins in the cells that are in very close contact with RNA molecules are thereby firmly bound to them. Scientists call this process “crosslinking.”
Over 100 sites discovered
In the next step, the RNAs and their binding proteins are fished out, purified and separated. The molecules are then ionized in a mass spectrometer, causing them to break apart into characteristic fragments. This allows the researchers to reconstruct the positions of the phosphate groups in the amino acid sequence of the RNA-binding proteins. The method becomes quantitative by comparing the percentage of phosphorylated with the corresponding free proteins (without the phosphate groups).
“Using this quantitative method, we identified more than 100 phosphorylation sites with regulatory potential on different RNA-binding proteins. This includes sites known to have regulatory function,” Vieira says. In collaboration with Professor Michael Gotthardt, an RBM20 expert who also works at the MDC, the researchers proved that phosphate groups on RBPs do actually have regulatory effects. They introduced RBM20 mutations into cell culture cells that could not be phosphorylated at certain sites or that do not carry phosphate groups but rather other chemical groups.
Alternative splicing is disrupted
We have identified phosphorylation sites that actually influence the regulation of RBM20.
The result was that alternative splicing was disrupted in the RBM20 mutants. RBM20 regulates this clever trick of nature. It allows a single gene to code for many variants of a protein – in this case, titin, which is so important for the heart. “The experiments with the mutants were a turning point for me. I was surprised by how clear the findings were, because usually a lot can go wrong in such experiments,” Viera emphasizes.
“This means we have identified phosphorylation sites that actually influence the regulation of RBM20,” Selbach explains. “It is the first indication that these sites are functionally important. But we cannot yet say whether they play a crucial role in heart muscle activity or in pathological changes in the muscle, such as those that occur in cardiomyopathies.”
In any case, the qRIC method appears to be the ideal complement to routine proteomic analyses, which identify all possible protein modifications that result from phosphorylation but cannot provide insight into their potential function.
Text: Catarina Pietschmann
Further information
- Researcher profile of Matthias Selbach: The protein detective
- Researcher profile of Michael Gotthardt: The perfectionist
- How to fill a heart
Literature
Carlos Henrique Vieira e Vieira et al. (2022): „Proteome-wide quantitative RNA interactome capture (qRIC) identifies phosphorylation sides with regulatory potential in RBM20“. Molecular Cell, DOI: 10.1016/j.molcel.2022.03.024
- Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC)
-
The Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC) is one of the world’s leading biomedical research institutions. Max Delbrück, a Berlin native, was a Nobel laureate and one of the founders of molecular biology. At the MDC’s locations in Berlin-Buch and Mitte, researchers from some 60 countries analyze the human system – investigating the biological foundations of life from its most elementary building blocks to systems-wide mechanisms. By understanding what regulates or disrupts the dynamic equilibrium in a cell, an organ, or the entire body, we can prevent diseases, diagnose them earlier, and stop their progression with tailored therapies. Patients should benefit as soon as possible from basic research discoveries. The MDC therefore supports spin-off creation and participates in collaborative networks. It works in close partnership with Charité – Universitätsmedizin Berlin in the jointly run Experimental and Clinical Research Center (ECRC), the Berlin Institute of Health (BIH) at Charité, and the German Center for Cardiovascular Research (DZHK). Founded in 1992, the MDC today employs 1,600 people and is funded 90 percent by the German federal government and 10 percent by the State of Berlin.