How pathogenic gene variants lead to heart failure
The molecular and cellular mechanisms that lead to heart failure in people with cardiomyopathy are determined by the specific gene variant that each patient carries, according to newly published research based on the first comprehensive single-cell analysis of cardiac cells from healthy and failing hearts.
The work, reported in the journal “Science”, was conducted by 53 scientists from six countries in North America, Europe, and Asia.
The study shows that cell type compositions and gene activation profiles change according to the genetic variants. The investigators say the findings can inform the design of targeted therapies that take into account each patient’s underlying gene defect responsible for their particular form of cardiomyopathy.
The team studied 880,000 single heart cells
In this illustration, the diseased heart is displayed as a piñata being hit by a DNA bat to signify the genetic impact of the variants that were studied. Patient cells are illustrated as confetti flying out of the heart and the colours recall the five main genotypes discussed in the paper.
Examining the genes activated in about 880,000 single cells from 61 failing hearts and 18 healthy donor hearts as reference was a complex endeavor which required an interdisciplinary team. The organs were procured by the Brigham and Woman’s Hospital in Boston, USA, University of Alberta in Canada, the Heart and Diabetes Center North Rhine-Westphalia in Bad Oeynhausen, Ruhr University Bochum in Germany and Imperial College London, UK.
Senior authors who spearheaded the project are Christine Seidman, professor of medicine and genetics at Harvard Medical School and a cardiologist at Brigham and Women’s Hospital; Jonathan Seidman, professor of genetics at Harvard Medical School; Norbert Hübner, professor of cardiovascular and metabolic sciences at the Max-Delbrück-Center for Molecular Medicine in the Helmholtz Association (MDC) and Charité – Universitätsmedizin Berlin as well as Dr. Gavin Oudit, University of Alberta, Canada; Professor Hendrik Milting, Heart and Diabetes Center NRW, Bad Oeynhausen, Ruhr University Bochum, Germany; Dr. Matthias Heinig, Helmholtz Munich, Germany; Dr. Michela Noseda of the National Heart and Lung Institute at Imperial College London, UK and Professor Sarah Teichmann, Wellcome Sanger Institute in Cambridge, UK. Joint first authors are Dr. Daniel Reichart (Harvard), Eric Lindberg and Dr. Henrike Maatz (both MDC).
A disease with many causes
The scientists focused on dilated cardiomyopathy (DCM), the most common form of heart failure that leads to heart transplantations. It involves an expansion (dilation) of walls of the heart chamber, especially in the left ventricle, the heart’s main pumping chamber. The muscles of the heart become weakened, compromising its ability to contract and pump blood, which ultimately leads to heart failure. The consortium studied tissues from patients with different genetic mutations that commonly lead to cardiomyopathies. These mutations occurred in proteins with different functions in the heart and the analyses indicate that these triggered different responses.
To our knowledge, this is the first such analysis conducted in cardiac tissue, and we hope this approach can be used to study other types of genetic heart diseases.
“We investigated pathogenic gene variants in heart tissue at the single-cell level, which allowed us to map precisely how specific pathogenic variants drive cardiac dysfunction,” says co-senior author Norbert Hübner. “To our knowledge, this is the first such analysis conducted in cardiac tissue, and we hope this approach can be used to study other types of genetic heart diseases.”
The scientists precisely characterized the various mutations in each of the hearts and compared them with each other, as well as with healthy hearts and with hearts in which the causes of dilation and dysfunction were unknown. Each cardiac cell type and the numerous subtypes were analyzed one by one, using single-cell sequencing methods. No lab on its own could deal with the massive amount of data generated, but close collaboration among specialists from different disciplines made it possible to assemble a coherent picture out of each individual piece of the puzzle. This study is also part of the efforts of the international Human Cell Atlas (HCA) consortium, which is aiming to map every cell type in the human body as a basis for both understanding human health and for diagnosing, monitoring, and treating disease.
“Only this level of resolution allows us to see that cardiomyopathies do not uniformly trigger the same pathological pathways,” says co-senior author Christine Seidman. “Rather, different mutations evoked specific and some shared responses that lead to heart failure. These genotype-specific responses point to therapeutic opportunities that may inform the development of precision-targeted interventions,” Seidman says.
Hyperactive connective tissue cells
The number of fibroblasts stays the same. But the existing cells become more active and produce more extracellular matrix, which fills in the space between the connective tissue cells.
“For example, we discovered that fibrosis – the abnormal growth of connective tissue – observed in DCM is not caused by an increased number of fibroblasts in the heart,” says Matthias Heinig, who carried out computational analyses. “The number of these cells stays the same. But the existing cells become more active and produce more extracellular matrix, which fills in the space between the connective tissue cells,” Eric Lindberg adds. Thus, rather than overproduction of fibrotic cells, the researchers observed only a shift in the proportion cell subtypes, marked by an increase in the number of fibroblasts specializing in the production of extracellular matrix.
The phenomenon was especially pronounced in the hearts of patients with a mutated RBM20 gene. This observation was also reflected in the patients’ medical histories.
“The phenomenon was especially pronounced in the hearts of patients with a mutated RBM20 gene,” Henrike Maatz explains. This observation was also reflected in the patients’ medical histories. On average, patients with this specific mutation suffered heart failure and needed a transplant much earlier than people with other genetic forms of DCM. Single-cell sequencing revealed a whole series of such genotype-specific differences in dilated hearts.
Specific patterns of change
The analysis also showed that in the hearts of people with arrhythmogenic cardiomyopathies (ACM) – those that cause dangerous heart rhythm disturbances – muscle cells are increasingly replaced by fat and connective tissue cells, particularly in the right ventricle. Although this form of cardiomyopathy can also be caused by a mutation in several genes the team focused its analysis on the gene for the protein plakophilin-2, or PKP2 for short. They compared cell signaling pathways from cells obtained from the right and left ventricles. The findings identify the cause behind increased cell-fat production in the cardiac muscle of people with this type of cardiomyopathy.
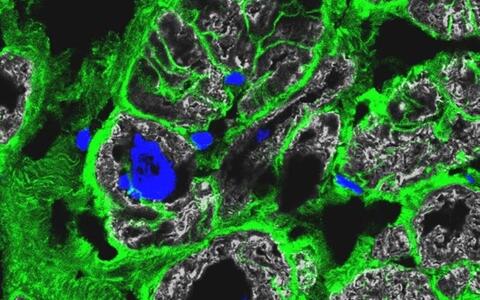
Immunofluorescence staining of diseased cardiac tissue. Cell boundaries are stained in green, nuclei are stained in blue, and heart muscle is stained in grey.
“The precise molecular signatures obtained for the highly specialized cells of the heart allowed us to predict cell-to-cell communication pathways,” Michela Noseda says. The team found that different genetic causes of cardiomyopathies were associated with specific aberrations of the cellular communication networks. “This is clear evidence of specified mechanisms driving the disease.”
Finally, the scientists used artificial intelligence to develop a model from all these data. Based on the specific patterns of molecular changes in the various cell types, the algorithm can predict with high degree of confidence which mutation is present – confirming that differences in gene and cell activation are associated with pathogenic variants of specific genes.
Biomarkers for targeted therapies
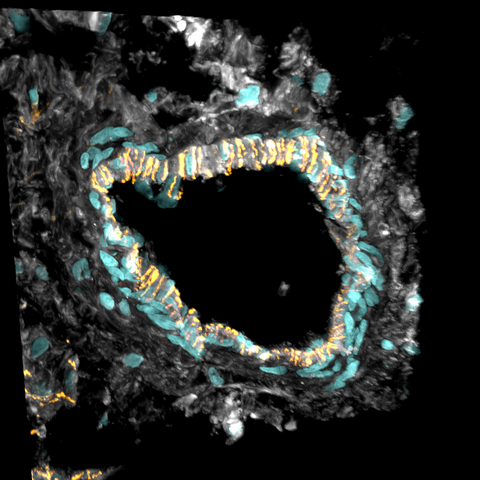
Cross section of an artery of the heart: The tissue was labelled using markers for specific cells. The cells lining the inside of blood vessels called endothelial cells are shown in orange. Cardiac muscle cells are in grey. Finally, in teal, are the cellular nuclei, the structures that contain the DANN – the genetic information – of each cell in the body.
The ultimate goal is to develop individualized therapies for heart disease the researchers said, because genotype-specific treatment could be more effective and with fewer side effects. The consortium has made all of its results available to the scientific community online. Seidman hopes that this resource propels studies by other groups to define new treatments that prevent heart failure, which today is an incurable disease.
“We investigated tissues of patients who needed a heart transplant; it was their last option,” Hendrik Milting says. “We hope that future pharmacological treatments will at least slow down the progression of the disease – and that the data from our study will help make this happen.”
Meanwhile, the research team has identified its next task. “The heart tissue that we studied came from people in the final stage of a disease,” Daniel Reichart, one of the first authors, says. “We are excited to see what changes we discover in earlier stages of disease, for example based on endomyocardial biopsies.” Perhaps biomarkers and pathways will be found that elucidate a very precise disease pathogenesis truly enabling personalized medicine, Gavin Oudit adds.
Further information
- All data from the study
- An in-depth atlas of the heart (press release)
Literature
Daniel Reichart, Eric L. Lindberg, Henrike Maatz et al. (2022): “Pathogenic variants damage cell compositions and single cell transcription in cardiomyopathies”. Science, DOI: 10.1126/science.abo1984
Photos to download
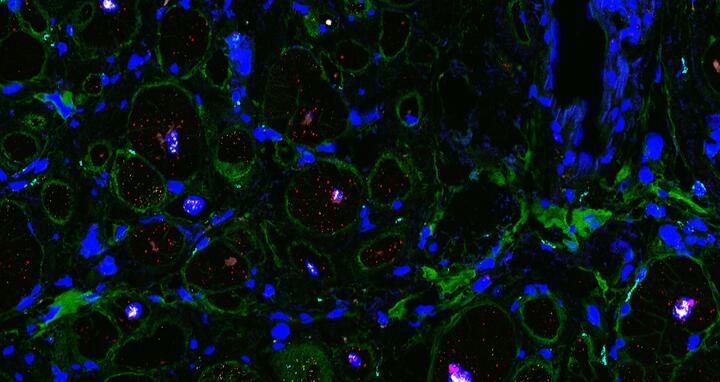
- Microscopy technologies help to dissect the cellular landscape of cardiac tissue. RNA molecules are labeled with fluorescent molecules and thereby help to identify cell-types. Cell boundaries and extracellular matrix are stained in green. Cell nuclei are stained in blue. © Eric Lindberg, MDC
- In this illustration, the diseased heart is displayed as a piñata being hit by a DNA bat to signify the genetic impact of the variants that were studied. Patient cells are illustrated as confetti flying out of the heart and the colours recall the five main genotypes discussed in the paper. © Dr. Eleonora Adami, MDC
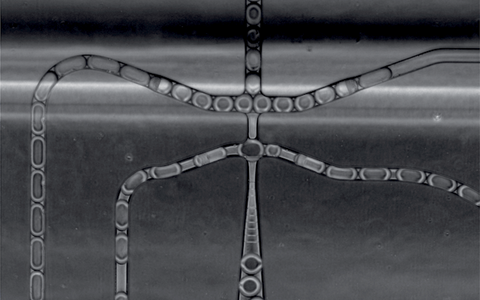
- For single cell sequencing, scientists first have to isolate the cell nuclei. When these nuclei pass over a microfluidic chip, they are packed into small aqueous droplets along with a barcode. © Eric Lindberg, MDC
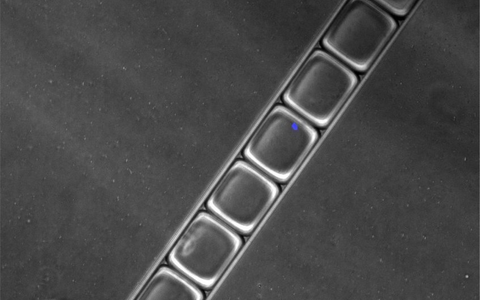
- In the droplets, the barcode is then coupled with the RNA from the nucleus - the transcripts that are generated when the genes are read from the DNA. This way, the RNA can be assigned to the individual cell nuclei after sequencing. The pictures show emulsion droplets, as they are formed after passing through the chip, with the cell nuclei stained in blue. © Eric Lindberg, MDC | Emulsion droplets in a network | Emulsion droplets in a row
- Machine learning algorithms enable a 2-D representation of the gene expression profiles. In this process, cells with similar gene expression patterns are spatially represented as neighbors. This allows different cell types to be distinguished from one another and – as shown here – cells of the human heart to be characterized. © Eric Lindberg, MDC
- Eric Lindberg at his work in the lab © Felix Petermann, MDC
- Immunofluorescence staining of diseased cardiac tissue. Cell boundaries are stained in green, nuclei are stained in blue, and heart muscle is stained in grey. © Anissa Viveiros and Dr. Gavin Oudit, University of Alberta
- Cross section of an artery of the heart: The tissue was labelled using markers for specific cells. The cells lining the inside of blood vessels called endothelial cells are shown in orange. Cardiac muscle cells are in grey. Finally, in teal, are the cellular nuclei, the structures that contain the DANN – the genetic information – of each cell in the body. © Sam Barnett and Antonio Manuel Almeida Miranda / Imperial College London
- An echocardiogram of a heart demonstrating dilation of the ventricle (increased chamber volume) and thinning of the ventricular wall, features that indicate reduced cardiac function that occurs in dilated cardiomyopathy. © Lech Paluszkiewicz, HDZ-NRW
Contacts
Professor Christine Seidman, M.D.
Director, CV Genetics Center at Brigham and Women’s Hospital
Harvard Medical School
cseidman@genetics.med.harvard.edu
Dennis Nealon
Associate Director, Media relations
Harvard Medical School
+1-508-494-6117
Dennis_nealon@hms.harvard.edu
Professor Norbert Hübner
Head of the Genetics and Genomics of Cardiovascular Diseases Group
Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC)
+49-30-9406-3512 (Secretariat)
nhuebner@mdc-berlin.de
Jana Schlütter
Editor, Communications Department
Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC)
+49-30-9406-2121
jana.schluetter@mdc-berlin.de or presse@mdc-berlin.de
- Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC)
-
The Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC) is one of the world’s leading biomedical research institutions. Max Delbrück, a Berlin native, was a Nobel laureate and one of the founders of molecular biology. At the MDC’s locations in Berlin-Buch and Mitte, researchers from some 60 countries analyze the human system – investigating the biological foundations of life from its most elementary building blocks to systems-wide mechanisms. By understanding what regulates or disrupts the dynamic equilibrium in a cell, an organ, or the entire body, we can prevent diseases, diagnose them earlier, and stop their progression with tailored therapies. Patients should benefit as soon as possible from basic research discoveries. The MDC therefore supports spin-off creation and participates in collaborative networks. It works in close partnership with Charité – Universitätsmedizin Berlin in the jointly run Experimental and Clinical Research Center (ECRC), the Berlin Institute of Health (BIH) at Charité, and the German Center for Cardiovascular Research (DZHK). Founded in 1992, the MDC today employs 1,600 people and is funded 90 percent by the German federal government and 10 percent by the State of Berlin.