E. Wanker Lab
Proteomics and Molecular Mechanisms of Neurodegenerative Diseases
Profile
The vinegar fly - Drosophila melanogaster by its zoological name - is a model organism that helps us understand how the disease affects nerve cells in the brain.
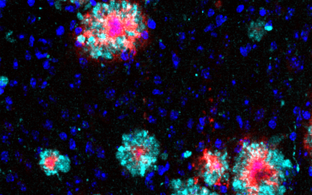
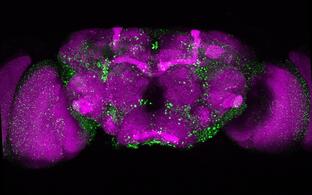
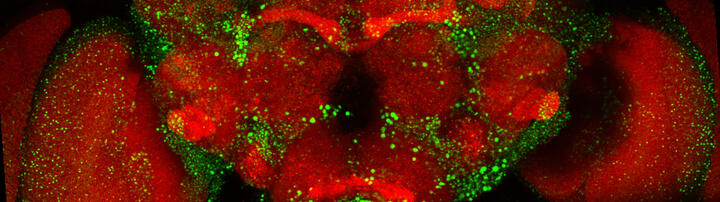
Confocal microscopy image of a Drosophila brain in which expression of huntingtin with an elongated polyglutamine sequence was turned on for 6 days and turned off for the 18 days that followed. Red: synapses present in all neuronal cells. Green: huntingtin aggregates that form and deposit in the brain.
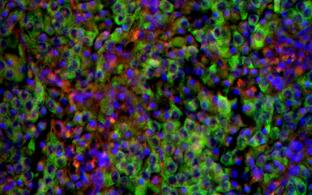
We have established HD fly models that enable the in vivo investigation of the huntingtin protein, its aggregation and seeding specifically in neurons. See: Anne Ast et al. “mHTT Seeding Activity: A Marker of Disease Progression and Neurotoxicity in Models of Huntington's Disease”. Molecular Cell. 2018 Sep 6;71(5):675-688.e6. https://doi.org/10.1016/j.molcel.2018.07.032.
More about our research:
Millions of people worldwide suffer from neurodegenerative disorders, like Alzheimer’s, Parkinson’s or Huntington’s disease. Most of these illnesses break out later in life. Correlated to the current demographic shift towards aging societies in many countries, the number of people affected with neurodegenerative diseases is growing.
Still, we do not understand exactly how neurodegenerative diseases develop. One of the characteristic features many neurodegenerative diseases share is the deposition of abnormally folded proteins in patient brains.
My group's research focusses on ‘Neuroproteomics’, the protein-based investigation of neurodegenerative diseases. We aim to elucidate the molecular principles by which proteins, sometimes abnormally folded, alone or in interaction, lead to cellular toxicity and neuronal dysfunction, causing neurodegeneration.
We pursue two main lines of investigation: Hypothesis-driven molecular studies of protein misfolding, aggregation and spreading, on the one hand, unbiased protein-protein interaction or ‘interactomics’ studies on the other. Click "Research" above to read about our current projects and fields of interest.
Team
Prof. Erich Wanker, PhD, DI
Principal Investigator
Orchid Wael Mostapha Ammar, MSc
Graduate student
Stephanie Beetz, MSc
Technician
Annett Böddrich, PhD, DBiol
Senior scientist, project manager
Annika Deckert, PhD, MSc
Postdoctoral scientist
Christian Hänig, DI
Computational scientist, IT, automation
Nancy Neuendorf, BTA
Senior technician
Roxane Maria Papawassiliou, MSc
Graduate student
Leonard Roth, MSc
Graduate student
Sigrid Schnögl, MPhil, MBA
Coordinator
Eduardo Silva Ramos, PhD, MSc
Postdoctoral scientist
Theo Wallenfang, MSc
Graduate student
Martina Zenkner, DI
Senior technician, lab manager
Research
In particular, we aim to elucidate the molecular principles by which abnormally folded proteins, their complexes and aggregates cause cellular toxicity and neuronal dysfunction. In our efforts to promote translation of basic research into benefits for patients, we identify and characterize modulators of protein misfolding cascades in disease (Ehrnhoefer et al., Nat Struct Mol Biol, 2008; Bieschke et al., Nat Chem Biol, 2011). We have previously demonstrated that expanded polyglutamine (polyQ) sequences trigger misfolding and aggregation of N-terminal huntingtin fragments in vitro and in vivo (Scherzinger et al., Cell, 1997; Davis et al., Cell, 1997). More recently, we have started new lines of translational research establishing methods to detect disease-relevant misfolded protein species in biosamples from models and patients. These investigations are directed at developing predictive disease markers that are a prerequisite for the clinical investigation of new disease-modifying therapies targeting neurodegeneration before symptoms of irreversible neuronal damage arise.
With our interactomics activities, we have developed an automated yeast two-hybrid (Y2H) system, which we used to generate a focused protein-protein interaction network for the huntingtin protein relevant to Huntington's disease (Goehler et al., Mol. Cell, 2004), as well as a large interaction map of the human proteome (Stelzl et al., Cell, 2005). More recently, we identified highly relevant interactions between the triple-A ATPase VCP/p97 and an adaptor protein that effects a fundamental structural change in VCP from a homohexamer to a heterooligomer with far-reaching functional implications (Arumughan et al. 2016). We are constantly developing more powerful methods for the identification and validation of protein-protein interactions, most recently LuTHy, a double readout luminescence-based technology for interactome mapping in mammalian cells (Trepte et al., Mol Syst Biol, 2018). A new resource for mapping the interactome of neurodegeneration has been available to researchers since 2020 (Haenig et al., Cell Rep., 2020):