Coscia Lab
Spatial Proteomics
Profile
Our translational research group develops and applies mass spectrometry (MS) based proteomics workflows for spatially-resolved systems biology. For this purpose, we use a combination of analytical tools including multi-level proteomics (expression-, interaction- phospho-proteomics) and functional cell biology assays.
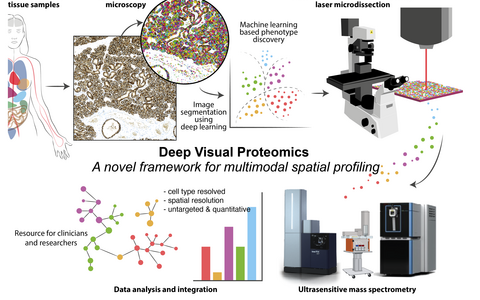
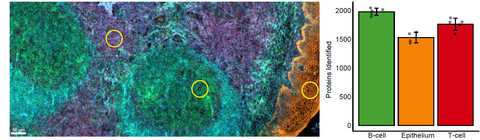
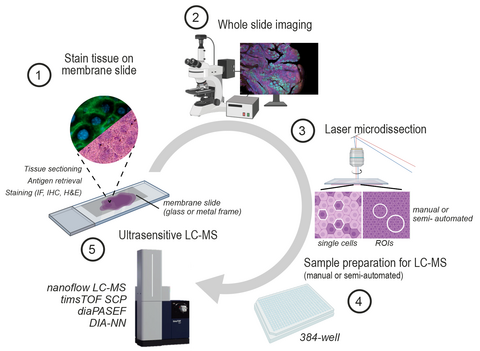
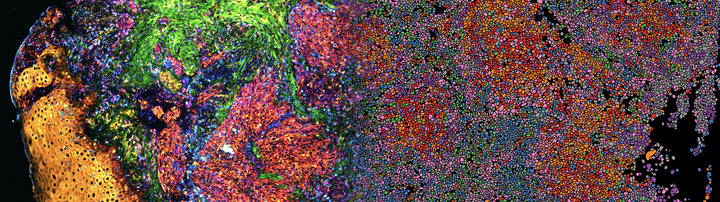
We have recently co-developed Deep Visual Proteomics, a first-of-its-kind concept that combines high-resolution imaging, AI-guided image analysis and ultra-sensitive MS-based proteomics. The method was highlighted as one of the Nature Methods' Method of the Year 2024. Applied to preclinical models and archived biobank specimens, this concept allows us to gain unprecedented insights into cellular proteome heterogeneity.
As part of the Berlin-centered Research Node ‘MSTARS’ (Multimodal clinical mass spectrometry to target treatment resistance), our MDC group closely collaborates with the Berlin Institute of Health, the Max Planck Institute for Molecular Genetics and the Humboldt University and the CHARITÉ. MSTARS, which is funded by the German Ministry of Science and Technology (BMBF) combines a broad range of complementary proteomic, metabolomic and imaging-based technologies with computational approaches for next-level patient care.
Our long-term vision is to decipher molecular and cellular disease pattern that can be used as predictive measures in precision oncology.
Team
Group Leader
- June 2021: Principal Investigator ‘Spatial Proteomics Group’
Max Delbrück Center for Molecular Medicine, Berlin, Germany. - 2017-2021: Post-Doc, Mann lab (Clinical Proteomics)
University of Copenhagen (NNF Center for Protein Research)
From 2019: Marie Skłodowska-Curie Postdoctoral Fellow - 2012- 2016: PhD candidate (Dr. rer. nat.), Mann lab
Max Planck Institute of Biochemistry, Munich, Germany - 2009-2011: Master of Science Molecular Biotechnology
TUM Munich, Germany.
Secretariat
Postdoctoral Researcher
Postdoctoral Researcher
PhD student
PhD student
PhD student
PhD student
PhD student
PhD student
Technical Assistant
Publications
News
Lab news
- Dec 2025: New publication! Our MCP paper is out where Melissa and team describe our optimized workflow for 100 spatial proteomes daily!
-
More info can be found here
- Nov 2025: Publication alert! :-) Our "Early Ovarian Cancer Study' is now published in Molecular Systems Biology!
Congratulations to the entire team, and specifically our first author Anuar!
More info here
- March 2025: Welcome Marcin!
Marcin has started his postdoctoral work in our lab focusing on blood cancers and tumor-macrophage interactions. We are very happy to have you in the group!
- Jan 2025: Welcome Daniela!
Daniela is starting her postdoctoral work in the lab applying spatial proteomics to women's health. Daniela's background is in placenta biology and preeclampsia research. We are very happy to have you in the group!
- Oct 2024: Welcome Lukas!! Our group is further growing and welcoming Lukas as new PhD student.
- May 2024: Big congrats to Sonja who is one of the winners of the Helmholtz 'Best Scientific Imaging Contest' !!!
-
- March 2024: Our second paper is out in Molecular & Cellular Proteomics!
We pioneered a robotic workflow for spatial tissue proteomics sample preparation based on the cellenONE. Congrats Anuar and Di for this shared success!
More info here: https://www.mcponline.org/article/S1535-9476(24)00040-9/fulltext
- March 2024: Big welcome to Rafael, who just started his postdoctoral work!
-
- Oct 2023: Our lab's first publication is out in Cell Systems!
- Highlights•Detailed protocol and guidelines for ultra-low-input spatial tissue proteomics•Single-cell MS-based proteomics data obtained from archival FFPE tissue•MS signal allows to normalize across tissue types and sample amounts•Cytokines and transcriptional regulators quantified in tonsil microregionsMore info here: https://www.cell.com/cell-systems/fulltext/S2405-4712(23)00287-9
-
- Sept 2023: Our lab was awarded an ERC Starting Grant!!!
- Please see more info about the project here: https://www.mdc-berlin.de/de/news/press/erc-starting-grant-fuer-fabian-… (English)and here: https://cms2.mdc-berlin.net/de/news/press/erc-starting-grant-fuer-fabia… (German)
-
- Aug 2023: What an achievement for Anuar winning the best short talk award at the Single-Cell Proteomics Conference in Vienna!
- Congratulations Anuar!
-
- July 2023: Melissa was awarded the MDC PhD fellowship!
- Congrats to Melissa Klingberg, who did her Master's thesis in our lab. She was now awarded a PhD fellowship position for the International PhD Program at the Max Delbrück Center! Congrats Melissa, we are very happy to have you in our group!
-
- May 2023: Our lab's first preprint is online! :-)
- Congrats to the entire team and our first author Anuar. We describe a scalable workflow for ultra-low input FFPE tissue proteomics. Our microscopy-guided proteomics approach shows that single-cell analysis from FFPE tissue is principally achievable! A great basis for many biomedical applications in the near future.
More infos here: https://www.biorxiv.org/content/10.1101/2023.05.13.540426v1
- St Nicholas Mass Spectrometry Symposium 2022
- Our group co-organized a Berlin networking event at the Charité, together with the Ralser, Demichev and Muelleder labs
The event was held on December 8th with over 150 participants from the Berlin area
- Oct 2022: New group picture
-
- First lab retreat 2022
- One full day of kayaking, swimming and relaxing. Exploring the beautiful nature around Berlin.
-